Key Points
GPIposCD56dim NK cells with an adaptive phenotype persist long-term in PNH patients.
Clonal tracking of adaptive NK cells in PNH patients suggests maintenance independent of HSPCs.
Abstract
Natural killer (NK) cells have long been considered short-lived effectors of innate immunity. However, recent animal models and human studies suggest that subsets of NK cells have adaptive features. We investigate clonal relationships of various NK-cell subsets, including the adaptive population, by taking advantage of naturally occurring X-linked somatic PIGA mutations in hematopoietic stem and progenitor cells (HSPCs) from patients with paroxysmal nocturnal hemoglobinuria (PNH). The affected HSPCs and their progeny lack expression of glycosylphosphatidylinositol (GPI) anchors on their cell surface, allowing quantification of PIGA-mutant (GPI-negative) HSPC-derived peripheral blood cell populations. The fraction of GPI-negative cells within the CD56dim NK cells was markedly lower than that of neutrophils and the CD56bright NK-cell compartments. This discrepancy was most prominent within the adaptive CD56dim NK-cell population lacking PLZF expression. The functional properties of these adaptive NK cells were similar in PNH patients and healthy individuals. Our findings support the existence of a long-lived, adaptive NK-cell population maintained independently from GPIposCD56dim.
Introduction
Natural killer (NK) cells are cytotoxic lymphocytes that lack rearranged antigen receptors yet possess an innate ability to kill virally infected or malignant cells.1 NK cells are phenotypically and functionally diverse, with responsiveness determined by expression of activating and inhibitory receptors and their interaction with target cell ligands.2 Recently, murine and non-human-primate models have uncovered adaptive NK-cell subsets able to maintain immunological memory to specific viruses or vaccines long-term,3-5 analogous to effector T cells. In humans, NK cells with biases in expression of specific cell surface receptors have been linked to cytomegalovirus (CMV) infection and reactivation.6-9 Most recently, absence of signaling proteins, including FcεRγ, SYK, and EAT-2, as well as downregulation of the transcription factor PLZF and a distinct epigenetic profile, was found to distinguish human canonical from potentially adaptive NK cells.10,11
Developmentally, NK cells are thought to originate from multipotent hematopoietic stem and progenitor cells (HSPCs) in the bone marrow and progress through phenotypic and functional maturation from CD56bright NK cells to cytotoxic CD56dim NK cells in lymph nodes or other tissues.12-14 Cell turnover studies have estimated a half-life of 14 days for circulating human NK cells and proliferation of 4% to 5% per day. These results were interpreted as evidence for ongoing production from immature progenitors, but a very rapid appearance of mature labeled cells in the blood, consistent with turnover of circulating cells, was also noted.15,16 Following transplantation of genetically barcoded autologous rhesus macaque HSPCs and lineage tracing, we found limited clonal overlap between the rhesus equivalent of CD56dim NK cells and other hematopoietic lineages, including CD56bright NK cells, suggesting independent production or maintenance of these NK cells.17
To ask whether ongoing production from HSPCs is required to maintain NK-cell homeostasis in humans, we took advantage of naturally occurring, somatic X-linked PIGA mutations in patients with paroxysmal nocturnal hemoglobinuria (PNH).18 PIGA encodes phosphatidylinositol N-acetylglucosaminyltransferase subunit A, which is required for the synthesis of glycosylphophatidylinositol (GPI) anchors. Acquired PIGA loss-of-function mutations occur in HSPCs of patients with PNH, resulting in production of hematopoietic cells lacking expression of GPI-anchored membrane proteins, including complement inhibitory proteins, leading to complement-mediated red cell lysis. PNH patients may exhibit stable mixed chimerism of GPI-positive (GPIpos) and GPI-negative (GPIneg) red cells and neutrophils for many years, reflecting ongoing output from both unmutated and PIGA mutated HSPCs, or eventually progress to virtually 100% GPIneg cells in these lineages. The extrinsic or intrinsic factors resulting in clonal expansion of HSPCs with PIGA mutations are poorly understood. Although total lymphocyte and NK-cell counts tend to be lower in PNH patients, immunodeficiencies have not been reported, and NK-cell function as well as overall distribution of NK-cell subsets appears to be preserved in these patients.19,20
Methods
Blood samples and cell preparation
All samples were collected under a protocol (04-H-0012) approved by the National Heart, Lung, and Blood Institute institutional review board, following written informed consent. Peripheral mononuclear cells and granulocytes were separated by density gradient centrifugation using LSM-lymphocyte separation medium (INC Pharmaceutical) according to manufacturer’s recommendations. Remaining red cells were lysed with ACK (ammonium-chloride-potassium) lysing buffer (Lonza) for 15 minutes at room temperature. Cells were resuspended in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, and 1000 U/mL penicillin-streptomycin (all Life Technologies) and either processed immediately for flow cytometry or cryopreserved in freezing media (RPMI1640, 40%FBS and 10% dimethyl sulfoxide).
Flow cytometry
Cell surface and intracellular staining of peripheral mononuclear cell markers was performed as previously described.10 Briefly, fresh or frozen samples were stained with FLAER (Alexa 488 proaerolysin variant) and antibodies or isotype controls (supplemental Table 1, available on the Blood Web site). Approximately 2 × 106 cells were stained for surface markers, fixable dead cell stain, and FLAER in fluorescence-activated cell sorting (FACS) buffer (PBS, 2% FBS and 2 mM EDTA). Cells were then fixed in 2% formaldehyde and permeabilized with 0.05% Triton X-100 followed by intracellular staining. For secondary staining, fluorochrome-labeled anti–mouse immunoglobulin M (IgM) or anti–rabbit IgG were used (supplemental Table 1). GPI expression on neutrophils and B lymphocytes was analyzed separately following the same staining procedure. Flow cytometry data were acquired on an LSR Fortessa-II cytometer (BD Biosciences), and the data were analyzed with FlowJo software (v9.9.3, FlowJo, LLC).
NK-cell functional assays
Functional studies were performed as previously described.10 Briefly, cryopreserved PBMCs from healthy volunteers or PNH patients were thawed and rested overnight in RMPI 1640 (Gibco) supplemented with 2 mM l-glutamine (Gibco) and 10% FCS (Sigma). Cells were cocultured in fresh medium with the mouse mastocytoma cell line P815 (ATCC) at a 2:1 ratio in the presence of 2 μg/mL purified anti-CD16 monoclonal antibody (3G8; BD Biosciences) or isotype control IgG1 (MOPC-21; BioLegend) as well as GolgiPlug and GolgiStop (both BD Biosciences). After 6 hours, cells were surface stained in FACS buffer as described followed by fixation in 2% formaldehyde (Polyscience), permeabilization with 0.05% Triton X-100 (Sigma), and staining of intracellular markers, including cytokines. Alternatively, cells were left untreated or stimulated with 10 ng/mL interleukin-12 (IL-12; Peprotech) and 100 ng/mL IL-18 (MBL) for 24 hours. After 18 hours, GolgiPlug was added to the culture. Cells were then surface stained, fixed, permeabilized, and stained intracellular as described above.
Barnes-Hut t-distributed stochastic neighbor embedding of multicolor flow cytometry data
As a dimensionality reduction technique for multicolor flow cytometry files, Barnes-Hut t-distributed stochastic neighbor embedding (SNE) was used.21 The raw flow cytometry data were compensated in FlowJo. Lymphocytes were gated via forward and side scatter and single cells on forward scatter height vs forward scatter area, followed by a gate on live CD3−CD4−CD14−CD19− cells. This resulted in a population that was further gated on CD56+ cells to define NK cells. Subsequently, data from 3000 gated NK cells (apart from donor 1, only having 590 events) were linearized and exported as comma separated values. In parallel, 1000 live CD14−CD19− cells from all individuals were exported (lymphocyte files). The data were then processed using R (version 3.30, 64 bit, The R Core Team). Robust normalization of all parameters in the NK-cell files was performed to the concatenated lymphocyte files from all individuals. The nine parameters (CD56, CD16, NKG2A, PLZF, FcεRγ, SYK, NKG2C, CD57, and CD2) were used to generate the SNE field. GPI-positive and GPI-negative cells and the distribution of these events were then plotted over the SNE field. For these analyses and all visualizations, the packages Rtsne, plyr, Hmisc, gplots, MASS, ggplot2, grid, and RColorBrewer were used.
Data and statistical analyzes
FlowJo (v9.9.3, FlowJo, LLC) was used for combinatorial Boolean gating. Pearson correlation coefficients were calculated using R. One-way ANOVA followed by multiple comparisons (Holm-Šídák) were performed using GraphPad Prism (v6.0).
Results and discussion
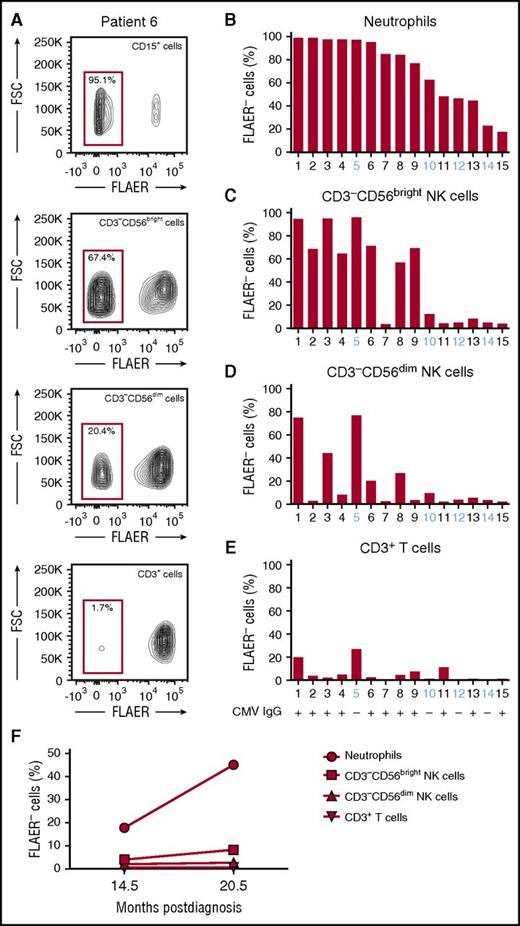
GPI expression can be quantified via flow cytometry using a labeled inactive variant of aerolysin (FLAER) that binds directly to GPI anchors.18 We analyzed GPI expression on neutrophils, total B and T cells, and NK-cell subsets from 15 PNH patients with GPIneg neutrophil chimerism ranging from 18% to 99% at a median time from diagnosis of 48.8 months (range, 3.5-158.4; Table 1; Figure 1A-B; supplemental Figures 1 and 2). The fraction of GPIneg neutrophils expanded over time in the majority of patients (supplemental Figure 2). The fraction of GPInegCD56bright NK cells (Figure 1C) correlated strongly with the fraction of GPIneg neutrophils (r = 0.84). In contrast, CD56dim NK cells had a significantly lower fraction of GPIneg cells that did not correlate with the fraction of GPIneg neutrophils (Figure 1D). Notably, T (Figure 1E) and B (supplemental Figure 1) cells were also predominantly GPIpos, reflecting peripherally maintained adult T- and B-cell homeostasis. Together, these results indicate that CD56dim NK-cell subsets may persist and propagate independently of CD56bright NK-cell precursors. This is further supported by longitudinal measurements of GPI expression in patient 15. While GPIneg cells increased by >100% in neutrophils and CD56bright NK cells in samples collected 6 months apart, the fraction of GPIneg in CD56dim cells remained unchanged (Figure 1F). In patients 1 and 3, with stable neutrophil GPIneg clone sizes >90% for many years, the markedly lower GPIneg chimerism in the CD56dim NK-cell population persisted during longitudinal follow-up (supplemental Table 2).
Patient characteristics
| ID . | Age (y) . | Sex . | Concurrent diagnosis . | CMV IgG/IgM . | Time GPIneg neutrophils > 1.0% (mo) . | Time GPIneg neutrophil >75% (mo) . | NK cell number (/µL)* . | GPIneg neutrophil at analysis (%) . |
|---|---|---|---|---|---|---|---|---|
| 1 | 23 | Male | AA | +/− | 78.2 | 78.2 | 41 | 99.2 |
| 2 | 43 | Male | AA | +/+ | 109 | 38.7 | 213 | 99.1 |
| 3 | 42 | Female | AA | +/− | 30.3 | 30.3 | 78 | 97.8 |
| 4 | 28 | Female | AA | +/− | 48.8 | 12.4 | NA | 97.6 |
| 5 | 36 | Female | AA | −/− | 131.6 | 100.8 | 75 | 97.3 |
| 6 | 33 | Male | AA | +/− | 61.4 | 36.6 | 91 | 95.2 |
| 7 | 60 | Female | AA/MDS | +/− | 15.1 | 5.2 | 141 | 85.1 |
| 8 | 67 | Female | AA | +/− | 75.1 | 0.0 | 55 | 84.4 |
| 9 | 29 | Male | AA | +/− | 17.7 | 11.7 | 184 | 77.1 |
| 10 | 59 | Male | AA | −/− | 38.6 | NR | 120 | 62.8 |
| 11 | 60 | Female | AA | +/− | 76 | NR | 71 | 48.3 |
| 12 | 61 | Male | AA | −/− | 17.2 | NR | 78 | 46.8 |
| 13 | 21 | Male | AA | +/− | 3.5 | NR | 198 | 44.7 |
| 14 | 32 | Female | AA | −/− | 158.4 | NR | 88 | 23.1 |
| 15 | 40 | Female | AA | +/− | 14.5 | NR | 116 | 17.8 |
| ID . | Age (y) . | Sex . | Concurrent diagnosis . | CMV IgG/IgM . | Time GPIneg neutrophils > 1.0% (mo) . | Time GPIneg neutrophil >75% (mo) . | NK cell number (/µL)* . | GPIneg neutrophil at analysis (%) . |
|---|---|---|---|---|---|---|---|---|
| 1 | 23 | Male | AA | +/− | 78.2 | 78.2 | 41 | 99.2 |
| 2 | 43 | Male | AA | +/+ | 109 | 38.7 | 213 | 99.1 |
| 3 | 42 | Female | AA | +/− | 30.3 | 30.3 | 78 | 97.8 |
| 4 | 28 | Female | AA | +/− | 48.8 | 12.4 | NA | 97.6 |
| 5 | 36 | Female | AA | −/− | 131.6 | 100.8 | 75 | 97.3 |
| 6 | 33 | Male | AA | +/− | 61.4 | 36.6 | 91 | 95.2 |
| 7 | 60 | Female | AA/MDS | +/− | 15.1 | 5.2 | 141 | 85.1 |
| 8 | 67 | Female | AA | +/− | 75.1 | 0.0 | 55 | 84.4 |
| 9 | 29 | Male | AA | +/− | 17.7 | 11.7 | 184 | 77.1 |
| 10 | 59 | Male | AA | −/− | 38.6 | NR | 120 | 62.8 |
| 11 | 60 | Female | AA | +/− | 76 | NR | 71 | 48.3 |
| 12 | 61 | Male | AA | −/− | 17.2 | NR | 78 | 46.8 |
| 13 | 21 | Male | AA | +/− | 3.5 | NR | 198 | 44.7 |
| 14 | 32 | Female | AA | −/− | 158.4 | NR | 88 | 23.1 |
| 15 | 40 | Female | AA | +/− | 14.5 | NR | 116 | 17.8 |
AA, aplastic anemia; MDS, myelodysplastic syndrome; NA, not available; NR, not reached.
Normal range, 126 to 729/µL.
Comparison of GPI presence in different blood cell lineages from PNH patients. (A) Representative FLAER staining analysis on blood cell populations from patient 6, showing FLAER− (GPIneg) and FLAER+ (GPIpos) neutrophils (CD15+), NK cells (CD3−CD19−CD14−CD56+), and T cells (CD3+). Separate analyses for CD56bright and CD56dim NK cells are shown. (B) Bar graph depicts the fraction of GPIneg granulocytes for each patient. Patient samples are ordered by contribution of GPIneg cells to the neutrophil compartment starting with largest clone size. (C-E) Bar graphs depict the fraction of GPIneg cells in other lineages, (C) CD56bright NK cells, (D) CD56dim NK cells, and (E) CD3+ T cells. Patient number and CMV status (IgG+ or IgG−) are indicated. (F) Longitudinal analysis of the fraction of GPIneg cells in each lineage over time in patient 15. The x-axis indicates the time postdiagnosis and the y-axis the fraction of GPIneg cells. FSC, forward scatter.
Comparison of GPI presence in different blood cell lineages from PNH patients. (A) Representative FLAER staining analysis on blood cell populations from patient 6, showing FLAER− (GPIneg) and FLAER+ (GPIpos) neutrophils (CD15+), NK cells (CD3−CD19−CD14−CD56+), and T cells (CD3+). Separate analyses for CD56bright and CD56dim NK cells are shown. (B) Bar graph depicts the fraction of GPIneg granulocytes for each patient. Patient samples are ordered by contribution of GPIneg cells to the neutrophil compartment starting with largest clone size. (C-E) Bar graphs depict the fraction of GPIneg cells in other lineages, (C) CD56bright NK cells, (D) CD56dim NK cells, and (E) CD3+ T cells. Patient number and CMV status (IgG+ or IgG−) are indicated. (F) Longitudinal analysis of the fraction of GPIneg cells in each lineage over time in patient 15. The x-axis indicates the time postdiagnosis and the y-axis the fraction of GPIneg cells. FSC, forward scatter.
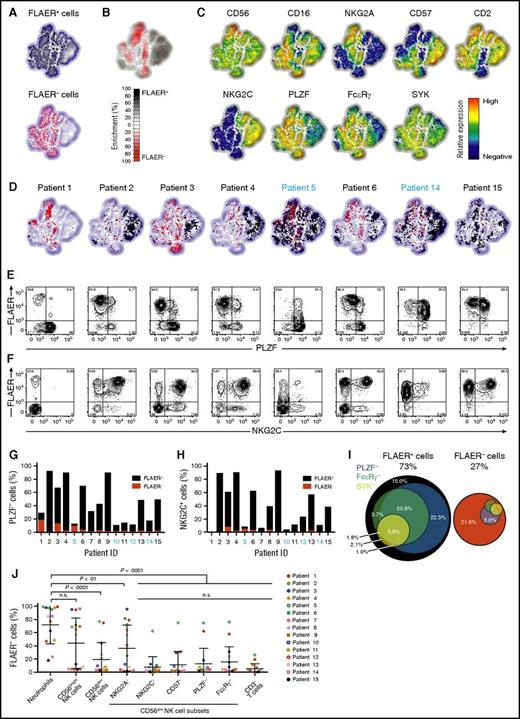
We next asked if GPI chimerism varied in CD56dim NK-cell subsets corresponding to adaptive versus canonical NK cells.8,10,22 Multivariate analyses of FACS data using nonlinear dimensionality reduction with stochastic neighbor-embedding approach (t-SNE) revealed strikingly different phenotypes of GPIpos and GPIneg NK cells (Figure 2A-C). Corresponding to adaptive NK cells, large CD56dimNKG2C+PLZF−FcεRγ+/−CD57+/− and smaller CD56dimNKG2C−PLZF−FcεRγ−CD57+ subsets were exclusively GPIpos (Figure 2B). Adaptive NK cells dominated in several patients (eg, 2 and 4), whereas adaptive GPIpos and canonical GPIneg NK cells coexisted in others (eg, 3 and 6; Figure 2D-F; supplemental Figure 3). Even in normal volunteers, the fraction of adaptive NK cells can vary markedly between individuals.10,11 These observations were confirmed through binary gating strategies, with PLZF–CD56dim and NKG2C+CD56dim NK cells overwhelmingly maintaining expression of GPI-linked proteins (Figure 2G-H). Overall, the majority of GPIpos NK cells displayed an adaptive NK-cell phenotype (Figure 2I). Conversely, GPIneg NK cells had significantly lower frequencies of NKG2C and CD57-expressing cells and mostly expressed PLZF and FcεRγ (Figure 2I-J).
NK cells with adaptive phenotype are largely GPI positive. PMBCs from 15 PNH patients were analyzed by flow cytometry. (A-D) Plots depict Barnes-Hut t-SNE analysis of 9-parametric data performed on gated CD56+ NK cells from all PNH patients. FLAER expression was not included as a parameter in the t-SNE analyses. (A) Distribution in the t-SNE field of GPIpos (FLAER+, top plot) and GPIneg (FLAER−, bottom plot) CD56+ NK cells from all patients. (B) Cumulative enrichment of GPIpos (FLAER+) and GPIneg (FLAER−) CD56+ NK cells from all patients according to the t-SNE field, as indicated. (C) Protein expression levels for single parameters in t-SNE field, as indicated. (D) Cell density in the t-SNE field for selected, individual patients, as indicated, with black and red dots indicating GPIpos (FLAER+) and GPIneg (FLAER−) CD56+ NK cells, respectively. Supplemental Figure 3 shows the individual t-SNE fields for all patients. (E-F) Flow plots depict (E) PLZF or (F) NKG2C vs FLAER expression on gated CD56dim NK cells in individual patients, as indicated. (G-H) Graphs indicate the frequency of (G) PLZF− and (H) NKG2C+ cells, also designating the proportion of which expressed FLAER or not, among CD56dim NK cells in individual patients, as indicated. (I) Venn diagrams depict the relative abundance of canonical and different adaptive CD56dim NK-cell subsets, as defined by lack of PLZF, FcεRγ , and SYK expression. Only samples with GPIneg neutrophils >75% are included (n = 9). (J) Analysis of the fractions of GPIneg cells in different NK cells subsets, neutrophils, and CD3+ T cells of 15 PNH patients. GPI expression was quantified in CD56dim NK cells either coexpressing NKG2A, NKG2C, and CD57 or lacking expression of PLZF or FcεRγ. The gating strategy defining positive and negative cells for each NK-cell marker, and expression profile for each patient, can be found in supplemental Figure 3. Each color depicts an individual patient. Full circles indicate CMV seropositive, whereas open circles indicate CMV seronegative. n.s., not significant, 1-way analysis of variance followed by multiple comparisons (Holm-Šídák).
NK cells with adaptive phenotype are largely GPI positive. PMBCs from 15 PNH patients were analyzed by flow cytometry. (A-D) Plots depict Barnes-Hut t-SNE analysis of 9-parametric data performed on gated CD56+ NK cells from all PNH patients. FLAER expression was not included as a parameter in the t-SNE analyses. (A) Distribution in the t-SNE field of GPIpos (FLAER+, top plot) and GPIneg (FLAER−, bottom plot) CD56+ NK cells from all patients. (B) Cumulative enrichment of GPIpos (FLAER+) and GPIneg (FLAER−) CD56+ NK cells from all patients according to the t-SNE field, as indicated. (C) Protein expression levels for single parameters in t-SNE field, as indicated. (D) Cell density in the t-SNE field for selected, individual patients, as indicated, with black and red dots indicating GPIpos (FLAER+) and GPIneg (FLAER−) CD56+ NK cells, respectively. Supplemental Figure 3 shows the individual t-SNE fields for all patients. (E-F) Flow plots depict (E) PLZF or (F) NKG2C vs FLAER expression on gated CD56dim NK cells in individual patients, as indicated. (G-H) Graphs indicate the frequency of (G) PLZF− and (H) NKG2C+ cells, also designating the proportion of which expressed FLAER or not, among CD56dim NK cells in individual patients, as indicated. (I) Venn diagrams depict the relative abundance of canonical and different adaptive CD56dim NK-cell subsets, as defined by lack of PLZF, FcεRγ , and SYK expression. Only samples with GPIneg neutrophils >75% are included (n = 9). (J) Analysis of the fractions of GPIneg cells in different NK cells subsets, neutrophils, and CD3+ T cells of 15 PNH patients. GPI expression was quantified in CD56dim NK cells either coexpressing NKG2A, NKG2C, and CD57 or lacking expression of PLZF or FcεRγ. The gating strategy defining positive and negative cells for each NK-cell marker, and expression profile for each patient, can be found in supplemental Figure 3. Each color depicts an individual patient. Full circles indicate CMV seropositive, whereas open circles indicate CMV seronegative. n.s., not significant, 1-way analysis of variance followed by multiple comparisons (Holm-Šídák).
To further characterize GPIposCD56dim NK cells, we performed combinatorial gating on this population using CD57, NKG2C, NKG2A, PLZF, and FcεRγ. Hierarchical clustering of patients based on the phenotype of GPIposCD56dim NK cells identified distinct sets of patients (supplemental Figure 4). One (cluster 1, n = 8) exhibited high abundance of adaptive CD56dimNKG2C+PLZF−FcεRγ−CD57+ cells and consisted of CMV-seropositive patients. The other (cluster 2, n = 7) included CMV-seronegative patients and CMV-seropositive without expansions of adaptive NK cells.
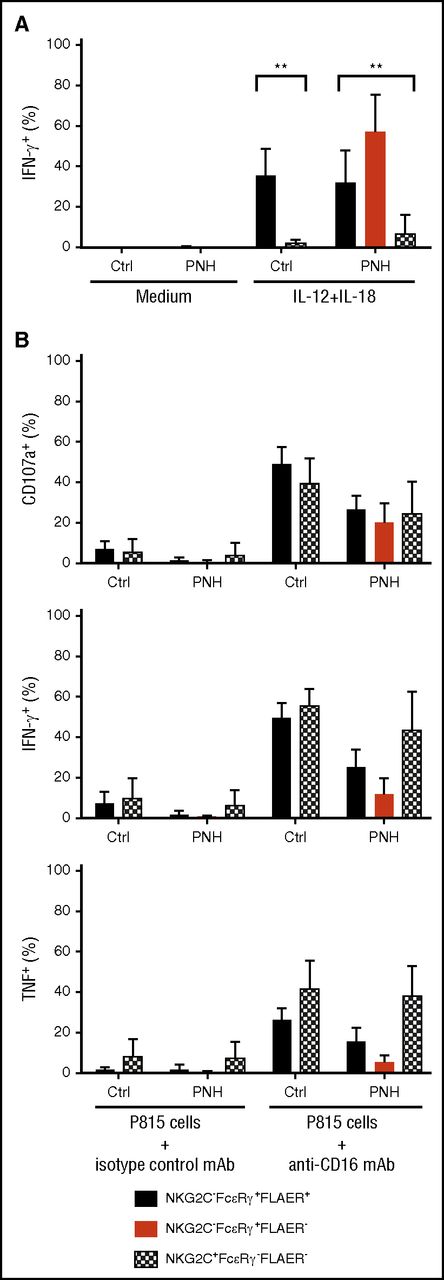
In order to determine whether the apparently long-lived GPIpos NK-cell subsets in PNH patients represented bona fide adaptive NK cells, we examined functional responses after cytokine stimulation or Fc receptor engagement in canonical (NKG2C−FcεRγ+) and adaptive (NKG2C+FcεRγ−) NK-cell subsets from PNH patients and healthy individuals. Humoral factors, such as IL-12 and IL-18, play central roles in stimulating cytotoxicity and cytokine production by human canonical NK cells, whereas adaptive NK cells display markedly diminished responses to these innate cytokines.10 In accordance with prior analysis of healthy volunteer samples, we found markedly reduced interferon γ (IFN-γ) production after IL-12 and IL-18 costimulation in adaptive GPIpos NK cells compared with residual canonical GPIpos NK cells from both PNH patients and normal controls (Figure 3A). Contrasting diminished responses to innate cytokines, adaptive NK cells retain degranulation and cytokine production following engagement of the low-affinity Fc receptor CD16, supporting the notion of adaptive NK cells as specialized to respond to target cell recognition of infected or potentially malignant cells.10 Activation of adaptive and canonical GPIpos NK cells in response to CD16 stimulation was comparable in PNH patients and healthy volunteer samples (Figure 3B), indicating that these long-lived GPIpos adaptive NK cells from PNH patients are functionally equivalent to previously-defined adaptive NK cells. Expression of cytokines IFN-γ and tumor necrosis factor appeared to be lower in GPIneg canonical NK cells than in residual GPIpos counterparts after anti-CD16 stimulation (Figure 3B). However, granule exocytosis following engagement of CD16 and IFN-γ production after IL-12 and IL-18 stimulation were comparable (Figure 3A-B), indicating that NK cells derived from GPIneg HSPC are generally functional. The very low number of GPIneg NK cells with an adaptive phenotype did not support functional evaluation.
Adaptive and canonical NK cells from PNH patients are functional. (A) PBMCs from CMV-seropositive healthy individuals (Ctrl, n = 5) or PNH patients (PNH, n = 10) were stimulated in the presence of IL-12 and IL-18. After 18 hours, canonical (NKG2C−FcεRγ+, filled bars, black for FLAER+ and red for FLAER− from the PNH patients) and adaptive (NKG2C+FcεRγ−, patterned, in PNH patients only FLAER− given lack of FLAER+ cells in this fraction) NK cells from controls and PNH patients were analyzed for intracellular IFN-γ expression. (B) Frequencies of adaptive and canonical NK cells showing degranulation (surface CD107a) and cytokine production (intracellular IFN-γ and tumor necrosis factor expression) after 6 hours of stimulation with P815 target cells and anti-CD16 monoclonal antibody (mAb). **P > .01, 1-way analysis of variance.
Adaptive and canonical NK cells from PNH patients are functional. (A) PBMCs from CMV-seropositive healthy individuals (Ctrl, n = 5) or PNH patients (PNH, n = 10) were stimulated in the presence of IL-12 and IL-18. After 18 hours, canonical (NKG2C−FcεRγ+, filled bars, black for FLAER+ and red for FLAER− from the PNH patients) and adaptive (NKG2C+FcεRγ−, patterned, in PNH patients only FLAER− given lack of FLAER+ cells in this fraction) NK cells from controls and PNH patients were analyzed for intracellular IFN-γ expression. (B) Frequencies of adaptive and canonical NK cells showing degranulation (surface CD107a) and cytokine production (intracellular IFN-γ and tumor necrosis factor expression) after 6 hours of stimulation with P815 target cells and anti-CD16 monoclonal antibody (mAb). **P > .01, 1-way analysis of variance.
Challenging the view of NK cells as short-lived HSPC-derived cells, clinical trials administering allogeneic NK cells and murine adoptive transfer studies have also provided evidence for in vivo proliferation and persistence of mature NK cells.23,24 Our analyses of PNH patients with major GPIneg neutrophil and erythrocyte clones present for up to 10 years reveal specific persistence of GPIpos adaptive NK cells. Our results suggest a long-lived source for adaptive CD56dim NK cells, independent of continuous production from HSPC. Notably, the only patient (#5) acquiring a high fraction of GPInegCD56dim NK cells was CMV seronegative, supporting the notion that specific viral exposure is required to confer longevity on mature NK cells. However, several CMV-seronegative PNH patients retained large populations of GPIposCD56dim NK cells with an adaptive phenotype, suggesting that that these patients harbored CMV in the absence of seroconversion or that alternative stimuli besides CMV may have resulted in the development of long-lived adaptive NK-cell clones.
A recent paper reported a higher GPIneg fraction of circulating CD56bright than CD56dim NK cells in PNH patients, which was hypothesized to be due to a specific defect in niche retention for GPInegCD56bright NK cells based on changes in chemotactic behavior of NK cells enzymatically deprived of GPI-anchored proteins in vitro.25 However, the authors did not include comparison with the overall HSPC PNH clone size in these patients or investigate NK cells in compartments outside the blood. Our functional and phenotypic studies did not uncover differences between control and PNH GPIpos NK cells.
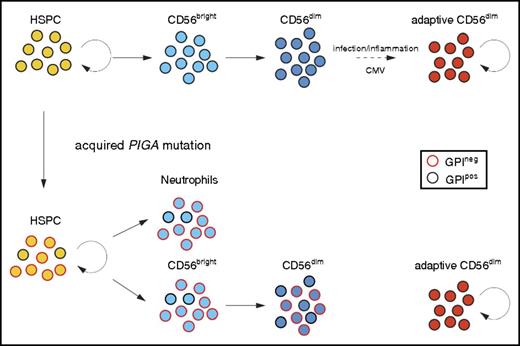
We believe our current data strongly support a separate, peripheral pathway for homeostatic maintenance of CD56dim NK cells with an adaptive phenotype, independent of ongoing production from HSPCs via CD56bright NK cells (Figure 4), analogous to long-lived and self-renewing memory T cells. Despite displaying complete dominance of GPIneg clones in the neutrophil lineage and in CD56bright NK cells over several years, a few of the analyzed PNH patients retained uniformly GPIpos adaptive NK cells. Further support for an independent self-renewal pathway for CD56dim NK cells comes from studies of patients with GATA2 mutation who develop progressive HSPC defects and lose CD56bright NK cells yet may preserve CD56dim NK cells.26 Recent data from Schlums et al demonstrate that residual CD56dim NK cells in these patients have an adaptive phenotype.27 These independent lines of evidence reveal that peripheral maintenance of adaptive NK cells is independent of HSPC or CD56bright precursor NK cells.
Somatic PIGA mutations in HSPCs reveals longevity of adaptive NK cells. Normal HSPCs give rise to CD56bright NK cells that further develop to canonical CD56dim NK cells. Adaptive, CD56dim NK cells derive from canonical CD56dim NK-cell precursors in response to environmental stimuli. Patients with an acquired PIGA mutation exhibit mixed chimerism of GPIpos and GPIneg myeloid cells (represented here by neutrophils), reflecting the ongoing clonal contribution of PIGA mutated self-renewing HSPC. CD56bright NK cells develop proportionally from GPIpos and GPIneg HSPC similar to myeloid cells. However, GPIneg cells are underrepresented in the CD56dim NK population, most prominently in CD56dim NK cells with adaptive phenotype. The persistence of GPIpos NK cells in PNH patients supports the notion of long-lived adaptive NK cells with self-renewal capacity similar to memory T cells.
Somatic PIGA mutations in HSPCs reveals longevity of adaptive NK cells. Normal HSPCs give rise to CD56bright NK cells that further develop to canonical CD56dim NK cells. Adaptive, CD56dim NK cells derive from canonical CD56dim NK-cell precursors in response to environmental stimuli. Patients with an acquired PIGA mutation exhibit mixed chimerism of GPIpos and GPIneg myeloid cells (represented here by neutrophils), reflecting the ongoing clonal contribution of PIGA mutated self-renewing HSPC. CD56bright NK cells develop proportionally from GPIpos and GPIneg HSPC similar to myeloid cells. However, GPIneg cells are underrepresented in the CD56dim NK population, most prominently in CD56dim NK cells with adaptive phenotype. The persistence of GPIpos NK cells in PNH patients supports the notion of long-lived adaptive NK cells with self-renewal capacity similar to memory T cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Flow Cytometry Core Facility and Keyvan Keyvanfar for flow cytometry support and the National Heart, Lung, and Blood Institute research nurses and clinical staff for sample procurement.
This project was supported by the intramural program of the National Institutes of Health National Heart, Lung, and Blood Institute, the European Research Council under the European Union’s Seventh Framework Programme (FP/2007-2013), ERC grant 311335, the Swedish Research Council, the Norwegian Research Council, the Swedish Foundation for Strategic Research, the Wallenberg Foundation, the Swedish Cancer Foundation, the Swedish Childhood Cancer Foundation, and the Stockholm County Council and Karolinska Institutet Center for Innovative Medicine (Y.T.B.).
Authorship
Contribution: M.A.F.C. collected and assembled data, analyzed and interpreted data, and wrote the manuscript; H.S. collected, assembled, analyzed, and interpreted data; C.W. collected and assembled data; J.T. and D.A.E. analyzed data; S.E.S. processed samples; D.M.T. and N.S.Y. provided study materials; Y.T.B. analyzed and interpreted data and wrote the manuscript; C.E.D. conceptualized the study, analyzed and interpreted data, and wrote the manuscript; T.W. conceptualized the study, collected, assembled, analyzed, and interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: C.E.D., N.S.Y., D.T., and T.W. received clinical research funding from Glaxo-Smith-Kline and Novartis. The remaining authors declare no competing financial interests.
Correspondence: Cynthia E. Dunbar, Hematology Branch, NHLBI/NIH, 10 Center Dr, Building 10/CRC, Room 4E-5132, Bethesda, MD 20892; e-mail: dunbarc@nhlbi.nih.gov.
References
Author notes
M.A.F.C. and H.S. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal