Abstract
The myelodysplastic syndromes (MDS) are initiated in hematopoietic stem cells (HSCs) that persist despite excellent clinical responses to DNA methyltransferase inhibitors (DNMTIs). Given the likely contributions of genetic and phenotypic heterogeneity to therapeutic resistance, we sought to characterize the response of MDS HSCs to DNMTIs at single cell resolution. We identified patients who received the DNMTI decitabine and either sustained hematologic improvement (n=2) or were non-responders (n=2). Bone marrow (BM) specimens were obtained at pre- and post-treatment time points, as well as from untreated MDS patients (n=2) and age matched controls (n=2). From these specimens, HSCs (Lin-CD34+CD38-CD90+CD45RA-) were FACS-purified and the Fluidigm C1 platform was used to capture 686 single cells, with subsequent RNA-sequencing (Illumina HiSeq2500, 100 bp paired-end). The data was of high quality with an average mapping rate of 80%.
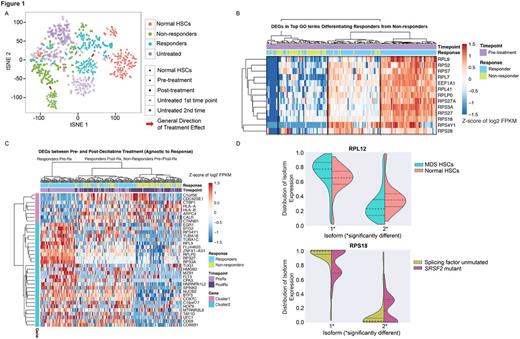
We used t-distributed Stochastic Neighbor Embedding (t-SNE) to visualize differences between cells from responders, non-responders, and age matched controls (Fig.1A). This revealed separation between responder and non-responder cells prior to treatment, with non-responders more distant from normal HSCs. Post-treatment samples revealed a shift in responder cells closer to normal HSCs, with non-responder cells remaining distant from normal HSCs. A small number of cells from responders and normal HSCs clustered away from their respective groups and closely with cells from non-responders, highlighting intratumoral transcriptional heterogeneity. We speculate that such cells represent a reservoir for eventual therapeutic resistance in responders. Comparing pre-treatment HSCs from responders and non-responders, we found 455 differentially expressed genes (DEGs, FDR<0.01). DEGs were enriched for gene ontology (GO) terms including mRNA catabolic processes, NMD, and translational elongation, with 12/13 DEGs in the top five GO terms being ribosomal proteins (RPs). Pre-treatment cells formed four clusters based on these genes (Fig.1B), with responders comprising the two highest expressing clusters and non-responders comprising the next to lowest expressing cluster. The lowest expressing cluster was a mixture of cells from responders and non-responders, again suggestive of a source for eventual therapeutic resistance in responders.
To assess the effect of decitabine on MDS HSCs agnostic to treatment response, we compared all pre-treatment MDS HSCs with all post-treatment MDS HSCs. We identified 45 DEGs (FDR <0.01) after filtering for high expression (FPKM >4) and removing genes that might reflect disease progression by excluding DEGs between early and late time points of untreated patients. Using these DEGs to cluster all pre- and post-treatment cells, the highest level of clustering separated a pre- and post-treatment cluster (Fig.1C). The pre-treatment cluster was primarily composed of pre-treatment cells from responders, with nearly all pre-treatment cells from non-responders falling in the post-treatment cluster, which also included post-treatment cells from both responders and non-responders. Thus MDS HSCs in responders occupy a unique transcriptional landscape prior to therapy.
As aberrant splicing may contribute to MDS pathogenesis, we developed an algorithm to evaluate differential isoform usage in single cells. Comparing MDS HSCs to normal HSCs, we identified 22 genes with differentially expressed isoforms, including seven RPs (Fig.1D). A comparison between SRSF2 mutant (n=3) and wild-type (n=4) patients identified 13 differentially expressed isoforms, including five RPs. This suggests that even MDS HSCs from patients lacking splicing factor mutations may exhibit aberrant splicing, and that these changes may differ from those found in patients with mutated splicing genes.
In conclusion, our studies of the transcriptional landscape of single MDS HSCs reveal RP gene signatures that represent novel biomarkers that distinguish MDS from age-matched controls and predict response to DNMTIs. Our studies also demonstrate significant intratumoral heterogeneity in gene expression that may underlie the biology of inevitable therapeutic resistance. Such heterogeneity includes differential isoform usage in both splicing factor mutant and wild type patients, suggesting that aberrant splicing may be a general feature of MDS.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal