Abstract
The initial clinical presentation of T-cell large granular lymphocytic leukemia (T-LGL) and myelodysplastic syndromes (MDS) can be similar, each characterized by unexplained peripheral cytopenias. However, these diseases are pathobiologically distinct and associated with stark differences in prognosis and therapy. T-LGL is a clonal lymphoid disorder defined by phenotypically abnormal cytotoxic T cells and an indolent clinical course, while MDS is a clonal disorder of hematopoietic stem cells defined by ineffective hematopoiesis, morphologic dysplasia, and an elevated risk of acute leukemia. Despite these differences, distinction between T-LGL and MDS can be challenging and misdiagnosis can significantly delay initiation of appropriate therapy. The recent identification of STAT3 mutations in LGL may facilitate this distinction: activating STAT3 mutations occur in 40-70% of T-LGL cases, primarily within the SH2 domain, but have not been reported in patients with MDS without concomitant T-LGL. STAT3 is included within our clinical next generation sequencing (NGS) panel, which is used to evaluate patients with known or suspected hematologic malignancies, primarily acute myeloid leukemia, MDS and myeloproliferative neoplasms, as well as various lymphocytic leukemias. We report the frequency and type of STAT3 mutations within our patient population and assess the impact of this information on diagnosis.
Between 1/1/2015 and 6/30/2016, 3414 samples (primarily peripheral blood (PB) or bone marrow (BM)) from 2530 unique patients evaluated at Brigham and Women's Hospital and/or Dana-Farber Cancer Institute underwent clinical NGS with a custom, 95-gene, amplicon-based panel (PMID: 27339098). Exons 2-17 and 21-23 of the STAT3 gene were analyzed in each sample using reference transcript 1 (NM_139276).
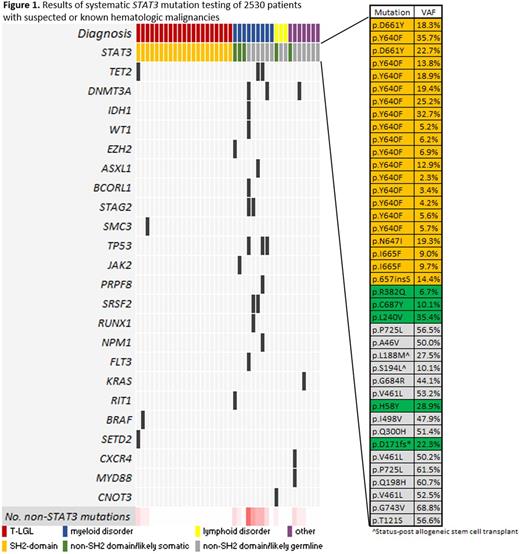
We identified 40 patients with 40 candidate STAT3 mutations (Figure 1). Based on domain localization, variant allele fraction, and population allele frequency, we classified these sequence variants as somatic SH2 domain mutations (n = 21), somatic non-SH2 domain mutations of unknown significance (n = 5) or germline variants (n = 14). Of the 21 patients with somatic SH2 domain mutations, 9 carried a prior diagnosis of T-LGL and 8 were concurrently diagnosed with T-LGL by conventional diagnostic criteria (clonal aberrant T cells in the setting of neutropenia, anemia, or lymphocytosis). The final 4 patients with STAT3 SH2 domain mutations were unexpected diagnoses of T-LGL. These 4 patients were initially referred from outside institutions for MDS based on the reported presence of unilineage erythroid dysplasia (n=2), unquantified ring sideroblasts (n=1), or pancytopenia with unspecified marrow findings (n=1). In 3 of these cases, the STAT3 mutation discovery prompted T-cell flow cytometric analysis of peripheral blood, which revealed an aberrant immunophenotype, and T-cell receptor gamma gene rearrangement studies, which were clonal; these tests are pending in the 4th case. BM evaluation was performed in 12 of 21 patients, including the 4 with suspected MDS; in all cases, the findings did not meet diagnostic criteria for MDS by expert hematopathology review and all showed a normal karyotype. Five additional cases demonstrated somatic non-SH2 domain STAT3 mutations of unknown pathobiologic significance: 3 myeloid neoplasms, 1 chronic lymphocytic leukemia, and 1 autoimmune hemolytic anemia. Additional non-STAT3 mutations were also frequently identified in tumors other than T-LGL.
Our experience demonstrates that STAT3 sequencing is a critical component of the evaluation of unexplained cytopenias, and identification of a mutation can clarify ambiguous phenotypes thus averting the consequences of misdiagnosis or diagnostic delay. Notably, several series have reported that MDS and T-LGL can infrequently occur concurrently and thus identification of a STAT3 mutation and a clonal T-LGL population does not exclude the possibility of concomitant MDS. In our cohort, however, no T-LGL patient with a STAT3 mutation demonstrated pathologic evidence of MDS and the majority (19 of 21) showed no other myeloid-associated somatic mutations. In addition, 5 cases in our cohort had likely somatic, non-SH2 domain STAT3 mutations in the context of disparate clinical scenarios, suggesting that these mutations may have a pathogenic role in other hematologic malignancies, a subject of future study.
DeAngelo:Ariad: Consultancy; Pfizer: Consultancy; Novartis: Consultancy; Amgen: Consultancy; Baxter: Consultancy; Incyte: Consultancy; Celgene: Consultancy. Stone:Celator: Consultancy; Jansen: Consultancy; ONO: Consultancy; Agios: Consultancy; Novartis: Consultancy; Pfizer: Consultancy; Celgene: Consultancy, Membership on an entity's Board of Directors or advisory committees; Juno Therapeutics: Consultancy; Amgen: Consultancy; Abbvie: Consultancy, Membership on an entity's Board of Directors or advisory committees; Karyopharm: Consultancy; Merck: Consultancy; Roche: Consultancy; Seattle Genetics: Consultancy; Sunesis Pharmaceuticals: Consultancy; Xenetic Biosciences: Consultancy. Lindsley:MedImmune: Research Funding; Takeda Pharmaceuticals: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.
This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal