Abstract
While the majority of leukemia cases occur in the absence of any known predisposing factor, there are germline mutations that significantly increase the risk of developing hematopoietic malignancies in childhood. Germline testing for the predisposition to myeloid malignancies is becoming more common with the recognition of multiple familial syndromes. In USA, Clinical Laboratory Improvement Amendments (CLIA) approved testing exists for mutations in RUNX1, GATA2, CEBPA and the other genes of inherited bone marrow failure syndromes. Meanwhile, GATA1 mutation is almost associated with Down syndrome and in acute myeloid leukemia (AML) with acquired trisomy 21. We experienced a case of infant AML with trisomy 21 and coexisting mutation of MUC16.
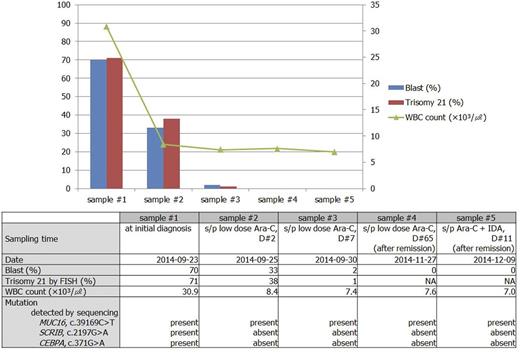
We experienced infant leukemia (FAB classification, AML M7, Acute megakaryoblasticleukemia) with trisomy 21 and coexisting MUC16, SCRIB, and CEBPA mutation in a 7-month-old boy. Peripheral blood (PB) was taken from the boy every day during hospitalization. Bone marrow (BM) examination was performed at initial diagnosis and after remission. All of PB and BM samples were collected with informed consent, and the study was reviewed and approved by the Institutional Review Board of Seoul National University College of Medicine. The selected five PB samples are demonstrated on Fig. 1. G-banding revealed trisomy 21 and fluorescent in situ hybridization (FISH) for chromosome enumeration 21 revealed 71% with trisomy 21 cells in his diagnosis. At first, we suspected transient abnormal myeloproliferative disease associated with mosaic Down syndrome and serially monitored the proportion of cells with trisomy 21 by FISH and the percentage of blast during hospitalization. Cells with trisomy 21 decreased as blast disappeared in PB during chemotherapy, suggesting trisomy 21 is acquired abnormality in leukemic cells.
We performed target gene sequencing of 359 genes related to the hematologic neoplasm, bone marrow failure syndrome, and cancer susceptibility to selected five PB. In addition, to search for the inherited predisposition gene to AML, we also performed whole genome sequencing (WGS) with BM specimen at initial diagnosis and after achieving remission. These next generation sequencing (NGS) with the Illumina HiSeq2500 platform revealed MUC16 (c.39169C>T) is the only significant mutation that persisted throughout from initial diagnosis to post-remission status. Mutations which were seen at initial diagnosis but disappeared after achieving remission were SCRIB (c.2197G>A, p.Arg733Trp) and CEBPA (c.371G>A, p.Ala124Val).
To investigate whether MUC16 (c.39169C>T) mutation is rare variant which can be detected in normal person, we screened MUC16 mutation in healthy control (n=365) and in patients with other hematologic diseases (adult myelodysplastic syndrome, n=155; adult aplastic anemia, n=57; adult myeloproliferative neoplasm, n=44; childhood myeloid neoplasm, n=26; inherited bone marrow failure syndrome, n=21; idiopathic eosinophilia, n=4; familiar hemophagocytic lymophohistiocytosis, n=10; congenital neutropenia, n=1) using allele-specific PCR. MUC16 (c.39169C>T) was found in none of them. Discovered site of MUC16 (c.39169C>T) was not reported in solid tumor as well, though the other sites of MUC16 were frequently reported. It is generally known that AML with trisomy 21 accompanies GATA1 mutation, but the infant AML in this study did not accompany GATA1 mutation, but rather, MUC16 mutation.
In this patient, GATA1 mutation was not found throughout hospital course. Instead, here we suggest that the constitutional MUC16 (c.39169C>T) mutation coexerts with acquired trisomy 21 in the development of infant AML and that MUC16 (c.39169C>T) mutation is a potential candidate gene for predisposition to AML.
Flow chart of PB blasts %, clonal cells % detected by FISH, mutated genes, and the corresponding clinical states.
Flow chart of PB blasts %, clonal cells % detected by FISH, mutated genes, and the corresponding clinical states.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal