Abstract
Background: Acute Myeloid Leukemia and Myelodysplasia are diseases associated with age and with genetic mutations driving evolution of clonal dominance. The earliest steps leading to genetic variation occur in a milieu of normal hematopoiesis by as a matter of course because genomic divergence is a fundamental element of biology. Organism speciation implicates genetic variation is the first step of evolution, followed by resource restriction pressure (a bottleneck). Such conditions create an environment where competitive advantages arise for genetically unique sub-species. Our work is based on the hypothesis that these rules also apply to the rise of pathologic clonal dominance. Consequentially, a low level of mutations in normal hematopoietic stem cells should be detectable and increase with age. Mutations can serve as lineage markers to allow the definition of the kinetics of competing myeloid sub-clones. Also, individual clones with higher rates of mutation might serve as a pre-condition for pathological clonal dominance and disease initiation.
Methods: Healthy 78 year old female myeloid progenitors were single cell sorted and cultured for 6 weeks in expansion media. Clones were harvested and typed apropos X-Chromosome Inactivation state (XCI state) as either "Xa" or "Xb" using a methylation specific polymerase chain reaction strategy after DNA was bisulfite modified. Remaining unmodified DNA was whole genome amplified and placed on an Affymetrix 6.0 Single Nucleotide Polymorphism (SNP) array (2.5 million SNP calls/array). Partek Genome Suite software interpreted raw SNP intensity signals as copy number variants (CNV) or loss of heterozygosity (LOH) calls using a stringent search strategy of a > 30 SNP markers, and an average variance of 0.5 (p<0.001). Mutations were compared between XCI states and within XCI states (i.e: individual Xa or Xb clones).
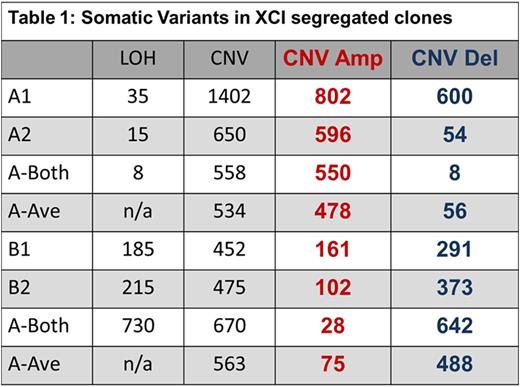
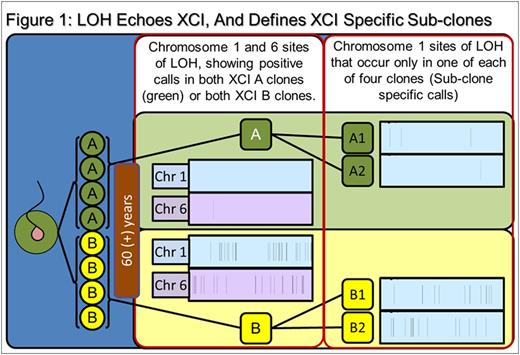
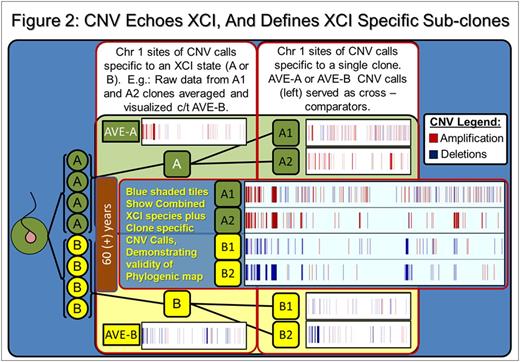
Results: SNP comparison between XCI defined phylogenetic lineages showed significant somatic LOH and CNV variance, as did comparison of clones within specific XCI states (e.g: between Xa clones). Mutations represent accumulated genetic drift over the 78.75 years since subject conception and are summarized in Table 1. Regarding call parameters, LOH analysis was unpaired, using Partek internal references. For CNV calls, subtractive comparison was made between related data sets. For example, raw Xa data from clones A1 and A2 were averaged, CNV calls were generated and compared to Xb averaged clone data (B1 and B2) to reveal specific XCI state variance (i.e.: A-Ave vs B-Ave). Individual clones were compared within the XCI state (i.e.: clone A1 compared directly to A2 to reveal unique A1 calls). An example of our data interpretation: 8 LOH and 558 CNV are specific to and shared among XCI state Xa clones (i.e.: A-both). Clone A1 had 35 specific, unique LOH and 1402 CNV calls, while clone A2 had 15 unique, specific LOH and 650 CNV calls. Xb clone data (B1, B2, B-both) are displayed in Table 1.
Conclusions: Recent studies reveal frequent mutations in the blood of healthy individuals; however within a person there a no model to identify normal myeloid sub-clones. Therefore we have no accurate data on stem cell kinetics or clonal evolution in healthy marrow. Herein we describe the feasibility of using SNP calls variation in un-diseased aged human myeloid cells to define stem cell specific progeny. To our knowledge, this is the first use of both inclusive (somatic mutations) and exclusive (XCI state) markers to develop a phylogenetic lineage map of normal aged marrow. Importantly, all clones had progenitors with an identical genetic background (the fertilized egg) at a single time point (conception). Therefore, this innovative approach allows for the somatic variation rate to be monitored in primary cells occupying the same organism environment as a function of age and stress events (i.e.: chemo, radiation, etc).
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal