Abstract
INTRODUCTION: Sickle cell disease (SCD) is characterized by the hemoglobin S (HbS) presence associated with other hemoglobin variants, as found in the hemoglobin SC disease (HbSC), or with globin chain synthesis defects, and sickle cell anemia (SCA) is the most severe form of SCD. Among clinical manifestations in SCD it has been highlight the stroke, which is the main cause of death among patients. Recent studies have associated the presence of gene polymorphisms with the stroke risk in SCA individuals. Several genetic markers have been previously associated with stroke risk, but many of the results remain controversial once they could not completely elucidate the effect of individuals' genetic heterogeneity in the stroke development. Thus, we sought to investigate genetic polymorphisms through genome-wide association among SCD patients with stroke risk.
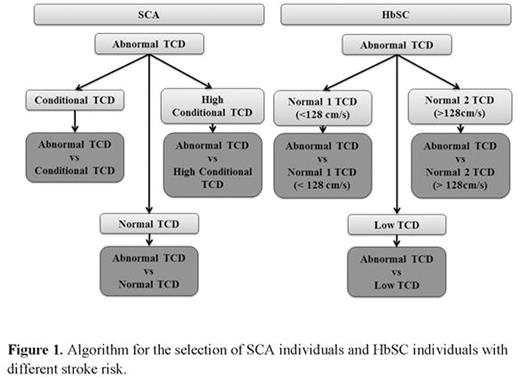
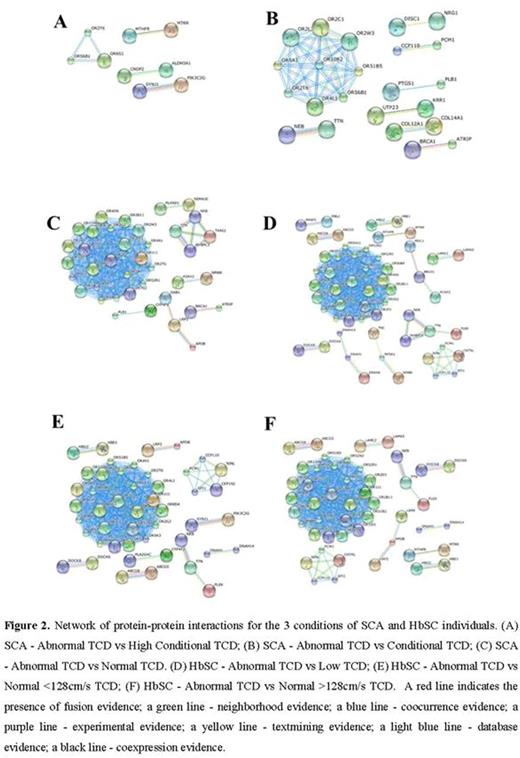
METHODS: We selected four SCA individuals and four HbSC individuals based on the transcranial Doppler (TCD) values in according to the velocities described by Adams et al and Deane et al respectively. Patients were matched by sex and age and are followed in the Pediatric Cerebrovascular Disease Outpatient Center at the Hospital Universitario Professor Edgard Santos of the Universidade Federal da Bahia. This study was approved by the Research Board of the Secretaria de Saúde do Estado da Bahia, and all experimental protocol were performed in accordance with the ethical standards of national research committee and with the Helsinki Declaration of 1975 and its later amendments. TCD was performed in all subjects included in the study, and the highest velocity found was assessed in the middle cerebral arteries and distal intracranial internal carotid. Genomic DNA was extracted from peripheral blood and the DNA concentration was evaluated and used for SNP analysis in an Illumina Human Omni5-4 v.1.1 BeadChip kit, and raw intensities were analyzed in GenomeStudio Software. We select SNPs showing different genotypes when individuals were pairwise compared according to TCD velocities such is showed in the Figure 1. We evaluate the SNPs using SNPnexus online tool and the UCSC, SIFT and POLYPHEN categories. The next step consisted of running these selected SNPs in the NHGRI-EBI Catalog of published genome-wide association studies with p-values < 1.0 x 10-5. Additionally, we used STRING database to construct a network of protein-protein interactions.
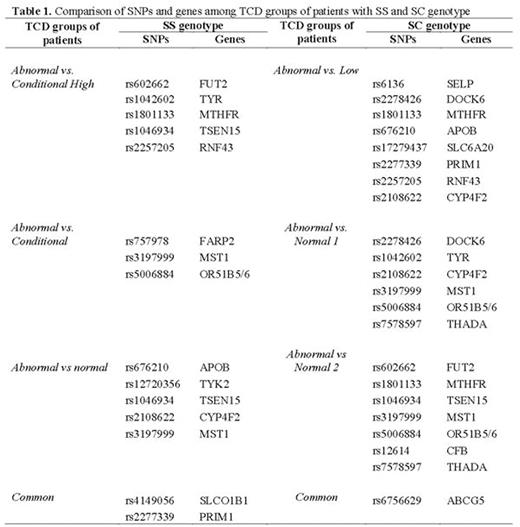
RESULTS: Using the UCSC category, we selected 3698, 3851 and 3528 unique SNPs in coding regions in non-synonymous for corresponding amino acid change for the three comparisons of SCA individuals respectively: abnormal TCD vs normal TCD, abnormal TCD vs conditional TCD and abnormal TCD vs conditional high TCD. Using the SIFT and POLYPHEN categories we selected the SNPs with a highly confident prediction to be damaging and SNPs predicted to be probably damaging, and we found 551, 505 and 598 unique SNPs and 424, 393 and 425 unique SNPs, in TCD-SCA individuals respectively. The next step was to select the common SNPs to perform Genome-wide association study (GWAS) classification. The same strategy used for TCD-SCA was performed to TCD-HbSC groups of individuals. We identified 3654, 3722 and 3847 as unique non-synonymous SNPs predicted by UCSC to be located in genomic coding regions respectively. Using the SIFT category 429, 447 and 418 unique SNPs were found in the TCD-HbSC groups of individuals respectively. PolyPhen classification was used and we found 517, 539 and 508 unique SNPs in the TCD-HbSC groups respectively. The next step was to perform GWAS classification. Results obtained utilizing the GWAS showed that each pair of individuals had a different number of SNPs identified (Table 1). The network of protein-protein interactions for the three conditions of SCA individuals and HbSC individuals are shown in the Figure 2.
CONCLUSIONS: We suggest that the DOCK6 rs2278426, TYR rs1042602, CYP4F2 rs2108622, MST1 rs3197999, OR51B5/6 rs5006884, THADA rs7578597, FUT2 rs602662, MTHFR rs1801133, TSEN15 rs1046934, CFB rs12614 and ABCG5 rs6756629SNPs may be candidate variants to be further investigated in HbSC individuals with high speed of cerebral blood flow. We suggest that the SLCO1B1 rs4149056, PRIM1 rs2277339, APOB rs676210, TYK2 rs12720356, TSEN15 rs1046934, CYP4F2 rs2108622 and MST1 rs3197999 as candidate SNPs to be further investigated in SCA with high cerebral blood flow velocities.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal