Abstract
Background: Extensive epidemiological data have demonstrated an exponential rise in the incidence of NHL associated with increasing age. The molecular etiology of this remains largely unknown. Additionally, recent data have suggested that older females with diffuse large B cell lymphoma (DLBCL) have improved survival when treated with rituximab-based therapy. We recently reported that there were significant global transcriptomic differences in DLBCL cases in the TCGA based on patient age and sex (Beheshti et al Cancer Inform. 2015). The hypotheses of the current studies were that age and sex-dependent circulating microRNA (miRNA) signatures exist and predict NHL initiation and progression. Leveraging a comprehensive systems biology approach in combination with a novel in vivo spontaneous DLBCL model, we sought to identify circulating miRNA signatures that drive initiation and progression.
Methods: We utilized a novel murine model for spontaneous DLBCL initiation (Smurf2-deficient mice with C57BL/6 background, Nat Commun. 2013) at two age groups: 2 and 15 months old. All Smurf2-deficient mice develop visible DLBCL starting at 15 months of age. Total miRNA was isolated from serum collected for all age groups for both Smurf2-deficient mice and wild-type C57BL/6 mice as controls. Using innovative technology with Droplet Digital and real time PCR (ddPCR), we quantified the 10 circulated miRNAs in the serum. Predictions on how miRNA signature impacts cancer risk were obtained using previously established methods (Beheshti et al. Cancer Res. 2015) by relating impact of miRNA on DLBCL and determined a cancer risk score based on fold-change differences compared with control. Additional effect of how miRNAs impact the transcriptome globally was completed via CluePedia Cytoscape plugin and functional significance on cancer was determined through Ingenuity Pathway Analysis (IPA).
Results: Using systems biology techniques, we identified 10 common circulating miRNAs that were significantly expressed in the spleen and bone marrow of Smurf2-deficient male and female mice at 2 months of age compared with wild-type mice. Furthermore, we predicted the impact it had on DLBCL initiation and progression from this miRNA signature (see Figure). For young Smurf2-deficient mice compared with wild-type mice, there was a predicted risk of cancer promotion due to the miRNA signature detection at 12 months prior to tumor formation. This miRNA signature was further modulated at older ages associated with the beginning of DLBCL formation. Additionally, this miRNA signature was more prominent in males and ongoing work is examining distinct miRNA associated with females. When determining the impact of the miRNA signature on DLBCL biology, we identified that these miRNAs have the most prominent impact on oncogenes, MYC and JUN with global impact on cancer related functions starting several months before actual histologic DLBCL formation.
Conclusions: It was determined that there is a key miRNA signature circulating throughout a host prior to the formation of a tumor starting at 2 months old which becomes modulated by age and tumor formation. This age based circulating miRNA signature can be used to predict DLBCL development at a young age before actual tumor formation. Furthermore, this can potentially be used as a simple biomarker at a young age to predict future lymphoma development and allow for advanced novel therapeutic strategies to prevent lymphomagenesis.
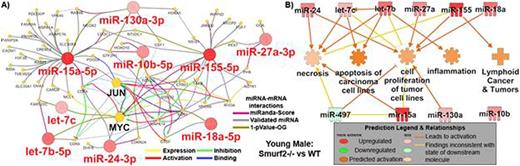
Impact of miRNA signature on transcriptional factors and cancer related function. A) For young males it was determined that the key miRNA signature has impact on oncogenes, MYC and JUN, along with other mRNAs determined using CluePedia Cytoscape plugin. The miRNA-mRNA interactions were determined from three different databases as indicated by the figure legend. B) Using Ingenuity Pathway Analysis (IPA) the functional impact of the miRNA signature is shown for young males comparing Smurf2-/- mice to wild-type (WT) mice.
Impact of miRNA signature on transcriptional factors and cancer related function. A) For young males it was determined that the key miRNA signature has impact on oncogenes, MYC and JUN, along with other mRNAs determined using CluePedia Cytoscape plugin. The miRNA-mRNA interactions were determined from three different databases as indicated by the figure legend. B) Using Ingenuity Pathway Analysis (IPA) the functional impact of the miRNA signature is shown for young males comparing Smurf2-/- mice to wild-type (WT) mice.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal