Abstract
Background: Pediatric acute lymphoblastic leukemia (ALL) is the most common pediatric malignancy and remains a leading cause of mortality and morbidity. Heterogeneity in the genetic and epigenetic landscape renders almost every patient genetically unique, making individualized medicine hard to achieve. Since genetic events are revealed by the expression and activation status of proteins, we hypothesized that these genetic events would coalesce into a finite number of protein expression signatures and that these could guide individualized therapy.
Methods:To determine relative protein expression patterns a custom Reverse Phase Protein Arrays (RPPA) with samples from 73 pediatric ALL patients and 10 normal CD34+ bone marrow samples were created and probed with 194 validated antibodies. As proteins interact with each other and act within networks, proteins were divided into 31 Protein Functional Groups (ProFnGrp) based on known associations from the literature. Progeny clustering was performed to determine the optimal number of protein clusters and principal component analysis was used to map global differences and similarities between protein clusters and normal CD34+ samples. Protein networks were constructed using literature associations and correlation within the data set. Associations between clinical features, outcomes and signatures were determined. Hierarchical clustering was performed on a compilation of all protein clusters into one binary matrix to identify recurrent protein expression signatures that comprised similar combinations of protein constellations. From this we constructed a list of proteins that were over or under expressed in each signature.
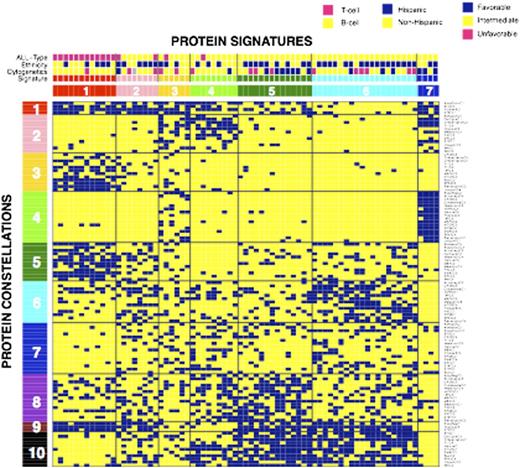
Results: Each ProFnGrp had 3 to 5 distinct expression clusters; at least one protein cluster was similar to the normal CD34+ samples in 23 of the 31 ProFnGrp and all had leukemia specific patterns. Protein expression levels were mapped onto the networks and showed different expression and activation states. Hierarchical clustering showed strong co-correlation between multiple groups of protein clusters from various ProFnGrp and suggested 10 protein constellations. Patients that expressed similar recurrent combinations of constellations formed 7 protein signatures (Figure). Most constellations and signatures were T- or B-cell specific, however 2 constellations showed overlap between the two diseases. Reanalysis limited to T-cell ALL revealed 3 protein signatures. Signature membership was correlated with overall risk stratification as determined by clinical features (P=0.001) as well as cytogenetics (P=0.017, Favorable risk with sig. 5 and 7, Intermediate risk with sig. 1 and 4) and absolute blast count (P=0.001). For both B- and T-cell ALL there were signatures that were significantly enriched for, and depleted of, patients of Hispanic ethnicity, suggesting that pathophysiological differences likely exist between non-Hispanic and some Hispanic ALL cases. Given the high CR and low relapse rates that resulted in a high overall survival in our cohort, signatures did not show significant correlation with clinical outcome. However, 3 of the 4 relapsing cases were in signature 6. The net level of expression and activation for each protein across each signature identified numerous proteins with significantly higher or lower expression, relative to normal CD34+ cells suggesting specific targets for combined targeted therapy.
Conclusion: Despite genetic heterogeneity ALL can be classified into a finite set of recurrent protein expression signatures. Signature membership correlated with risk stratification and cytogenetics. The identification of signatures associated with Hispanic ethnicity suggests that pathophysiology, rather than socioeconomic factors may underlie the inferior outcome of Hispanic patients. The net protein expression and activation status within each signature suggest targets for directed combinatorial inhibition or replacement to enable individualized therapy.
Hierarchical clustering based on binary ProFnGrp cluster membership. Each vertical patient column consists of 31 out of the 114 protein clusters. The suggested number of protein expression signatures is 7 together with 10 protein constellations. Blue squares indicate positive cluster membership.
Hierarchical clustering based on binary ProFnGrp cluster membership. Each vertical patient column consists of 31 out of the 114 protein clusters. The suggested number of protein expression signatures is 7 together with 10 protein constellations. Blue squares indicate positive cluster membership.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal