Abstract
Fate commitment of hematopoietic progenitor cells (HPCs) is thought to occur through a series of hierarchical fate choices, established over the past four decades through live cell tracking, in vitro colony-forming assays and transplantation of defined sub-populations of HPCs. Depictions of the HPC hierarchy invoke a tree structure, with gradual lineage-restriction at branch points, although the precise tree remains controversial. A second controversy relates to the nature of undifferentiated MPPs. MPPs have been suggested to express conflicting lineage-restricted programs indicating multi-lineage priming, or to host distinct cell sub-populations with intrinsic biases, or alternatively to form an entirely na•ve state. We asked whether the molecular state of cells seen by RNA profiling of thousands of single HPCs could resolve these controversies, by correctly predicting the known fates of sub-populations of HPCs reported over the past two decades, defining MPP heterogeneity, and defining the topology of HPC fate commitment.
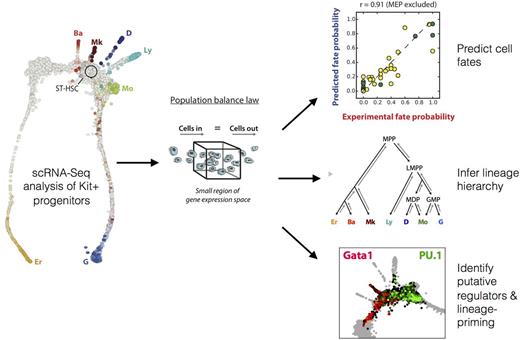
Predicting the future behavior of cells from high-dimensional snapshots of their current state is an unsolved problem. Until now, snapshots of single cell molecular states have been used to order events in cell differentiation, cell cycle, and perturbation response by methods that fit cell states to a curve or a tree, but these approaches tend to be suggestive rather than predictive. Here, we invoke a conservation law Ð commonly known as "population balance" Ð to formally predict differentiation fates from single cell molecular profiles, using an approach that is asymptotically exact given certain assumptions about the differentiation process. Application of the conservation law to single cell RNA profiles required developing novel mathematical results, leading to an algorithm termed Population Balance Analysis (PBA).
We apply PBA to bone marrow hematopoiesis, and recover the structure of the hematopoietic progenitor cell (HPC) population as it differentiates into seven lineages. The inferred fate choices reconcile fate-potential assays from the past two decades, and suggest a unified molecular definition of the hematopoietic hierarchy. In contrast to the canonical hierarchy, we predict that HPCs do not form a strict tree, and that MPPs show evidence of multi-lineage priming with simultaneous low-level gene expression of conflicting differentiation programs. In normal hematopoiesis, we predict a novel erythroid-basophil progenitor cell state, whereas erythroid/megakaryocyte fates become coupled in stress. We predict several novel regulators of cell fate at choice points in hematopoiesis. Overall, this work reconciles novel RNA-Seqdata with gold standard functional fate assays; it predicts novel regulators of hematopoiesis; and it establishes a predictive analytical method for dissecting complex differentiation hierarchies from single cell molecular profiles.
Klein:OneCell Bio: Equity Ownership, Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal