Abstract
According to standard practice, red blood cells stored for transfusion are kept at 4°C for up to 42 days. Studies on these RBC units therefore require over a month of experimental work. At higher temperatures, dynamics proceed at an increased rate, creating the potential for faster experiments. In this study, twelve RBC units were stored in SAGM at 4°C, 13°C, 22°C, and 37°C and profiled using quantitative metabolomics throughout storage in order to determine whether the metabolic RBC response is similar at higher temperatures.
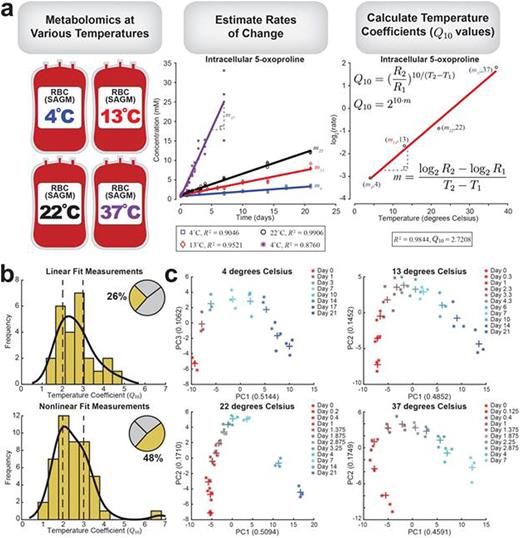
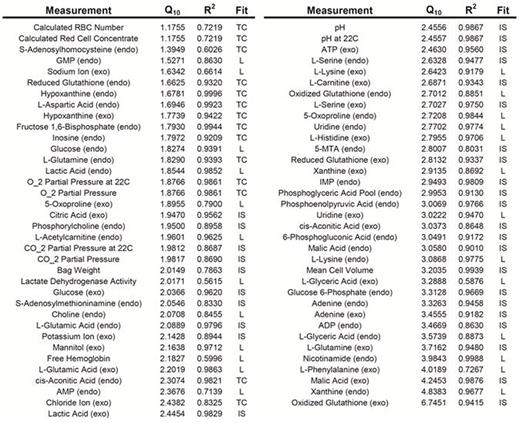
In order to obtain a rate for each measurement, the profiles were fit as either linear or nonlinear. A quarter of the data (26%) was classified as linear, while approximately half of the measurements (48%) were classified and fit as nonlinear. The remaining 26% of the measurements were either noisy or exhibited no clear trend and were excluded from further calculations. Once a rate was calculated for each measurement, it was plotted on a log2(rate) vs. temperature plot. Linear regression was used to calculate the slope, from which the temperature coefficient (Q10) was calculated (Fig. 1a). The resulting distribution of temperature coefficients shows consistent values in the 2 to 3 range, the typical estimate for biological systems (Fig. 1b); the median Q10 of the linear fits was 2.64 (mean = 2.64), and the median Q10 of the nonlinear fits was 2.44 (mean = 2.52). A complete list of all linear and nonlinear fitted measurements with corresponding Q10 values is provided in Table 1.
Principal component analysis (PCA) was performed on the data, yielding consistent baseline behavior in three components: one component was highly correlated with time (~50% contribution), one bimodal phase correlated with oxidative measurements (~10% contribution), and one component represented bag variance (~5-10% contribution). The component that was correlated with oxidative measurements and 2,3-diphosphoglyceric acid depletion displays the same qualitative behavior when plotted against the time component at all four temperatures (Fig. 1c). As temperature increased, the trend was accelerated.
Looking at specific pathway behavior, we observed a significant accumulation of metabolites in the nucleotide salvage pathway (e.g., xanthine, hypoxanthine, uridine) at higher temperatures, while concentrations were fairly steady at low temperatures. The same behavior was observed for intracellular citric acid. Several metabolites proved to be outliers in scaling behavior, notably intracellular xanthine and extracellular oxidized glutathione, which had the highest calculated Q10 values (4.84 and 6.75, respectively).
Metabolite profiling for RBCs in storage at different temperatures show that the rate of change of metabolite profiles approximately double for every 10°C of change in storage temperature. The increased metabolism causes a pronounced accumulation or depletion of certain metabolites at 22°C and 37°C, while the behavior at 13°C was similar to that at 4°C. These results indicate that for metabolic studies, storage at temperatures as high as 13°C provides a similar but accelerated system that could reduce the time required to complete experiments.
Overview of data. (a) Workflow for data analysis. Metabolomics data was collected at 4 different temperatures (4, 13, 22, and 37 degrees Celsius) across multiple time points. The concentrations were plotted against time, and the rates of change were estimated. For measurements that were fit as either linear or nonlinear profiles, log2(rate) was plotted against temperature in order to calculate the temperature coefficient. (b) Histograms show the distribution of temperature coefficients for the linear and nonlinear measurements separately; vertical dashed lines are at Q10 = 2 and 3, the typical estimated range for biological systems. The inset pie chart shows the percentage of measurements that used a linear and nonlinear fit; the remaining portion (26%) represents measurements that were either noisy or did not show a clear trend. (c) PCA plots show the same but accelerated trend at the four different temperatures; the Day 7 measurement is colored the same (cyan) in each plot for reference. + indicates the mean of three replicates at a particular time point.
Overview of data. (a) Workflow for data analysis. Metabolomics data was collected at 4 different temperatures (4, 13, 22, and 37 degrees Celsius) across multiple time points. The concentrations were plotted against time, and the rates of change were estimated. For measurements that were fit as either linear or nonlinear profiles, log2(rate) was plotted against temperature in order to calculate the temperature coefficient. (b) Histograms show the distribution of temperature coefficients for the linear and nonlinear measurements separately; vertical dashed lines are at Q10 = 2 and 3, the typical estimated range for biological systems. The inset pie chart shows the percentage of measurements that used a linear and nonlinear fit; the remaining portion (26%) represents measurements that were either noisy or did not show a clear trend. (c) PCA plots show the same but accelerated trend at the four different temperatures; the Day 7 measurement is colored the same (cyan) in each plot for reference. + indicates the mean of three replicates at a particular time point.
Q10 values (with corresponding R2) for all linear and nonlinear fit measurements. Abbreviations: L, linear fit; TC, time constant (nonlinear fit); IS, initial slope (nonlinear fit).
Q10 values (with corresponding R2) for all linear and nonlinear fit measurements. Abbreviations: L, linear fit; TC, time constant (nonlinear fit); IS, initial slope (nonlinear fit).
Bordbar:Sinopia Biosciences: Employment, Patents & Royalties. Palsson:Sinopia Biosciences: Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal