Abstract
Background:After the positive results of the Quiredex trial suggesting a benefit in time to progression (TTP) and overall survival for high risk SMM patients treated with lenalidomide and low dose dexamethasone, significant attention has been paid to this patient-population as potential candidates for enrollment in clinical trials exploring early treatment intervention. This requires accurate patient selection, and the Pethema criteria to identify high risk SMM patients [those with >95% clonal plasma cells (PCs) within the total bone marrow (BM) PC compartment plus immune paresis] has raised some criticism due to the perception that MFC immunophenotyping is difficult to standardize. Although in spite of such limitations, the prognostic value of MFC in SMM has been reproduced by other groups, novel technological developments that could help standardizing MFC would be mostly welcomed.
Aim: To develop a novel automated MFC classification algorithm to identify SMM patients at different risk of progression.
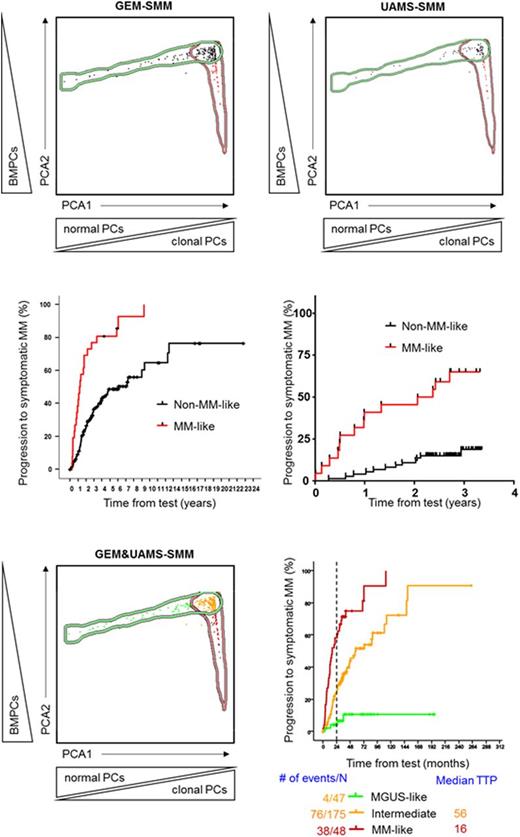
Methods: A total of 270 SMM patients from two independent series were included in the present study; one from Spain (n=174) and another from Little Rock, Arkansas (n=96), hereafter named as GEM-SMM and UAMS-SMM. Median follow-up for each series was of 66 and 31 months, respectively. First, we started by determining the relative frequency of BMPCs plus the percentage of clonal and normal PCs within the whole BMPC compartment for each SMM patient. Since our aim was to discriminate within the total SMM population, those that phenotypically resemble (and clinically behave) like MGUS and like MM, each SMM patient was compared with a dataset containing information on the same three parameters from a total of 1419 patients, including 497 MGUS and 922 symptomatic MM. Using new EuroFlow software tools, principal component analysis (PCA) using those three parameters was then performed and shown by the automatic population separator (APS; PCA1 vs. PCA2) graphical representation of the Infinicyt software (Cytognos). On the basis of the APS view, MGUS and symptomatic MM patients were simultaneously grouped and defined by 1.5 standard deviation median borders, whereas for SMM patients each one of them became represented by a dot, and the first (x-axis) and second (y-axis) principal components were used to produce a bi-dimensional representation of patient profiles. Ultimately, each SMM case was plotted against the dataset, and the Infinicyt software automatically classified the patient as MGUS-, intermediate- or MM-like.
Results: We started by investigating the prognostic impact of the novel and automated MFC algorithm in each of the two independent series. Among GEM-SMM patients, those classified as MM-like (n=26/174; 15%) showed significantly shorter median TTP than the remaining SMM cases (medians of 14 vs 73 months; HR: 3.5, P<.0001). Similarly, UAMS-SMM patients being classified as MM-like (n=22/96; 23%) also showed significantly inferior TTP as compared to the remaining SMM cases (median of 27 months vs NR; HR: 5.5, P<.0001). After validating the novel MFC algorithm in the two independent series, we merged them in order to enhance risk classification of SMM patients. Our results showed the heterogeneity that underlies beneath SMM, with approximately two-thirds of the cases (n=175; 65%) being plotted in the intermediate area between MGUS and MM, 47 (17%) patients being classified as MGUS-like and 48 (18%) cases being classified as MM-like. This MFC-based classification translated into markedly different TTP into MM: median NR and 4% risk of progression at 2-years for MGUS-like, median of 57 months and 25% risk of progression for intermediate, and median TTP of 16 months and 58% risk of progression at 2-years for MM-like SMM patients (P<.0001). Noteworthy, when we restricted the analysis to the GEM-SMM series due to the shorter follow-up of the UAMS-SMM cohort, risk of progression at 2-years for MM-like SMM patients increased to 69%.
Conclusions: We developed a MFC computerized classification algorithm that as a single biomarker (without immune paresis), identifies SMM patients with high-risk of transformation to active MM. Importantly, this classification algorithm can be fully standardized (through automatized analysis of flow data) and as shown here, has the potential to be shared across different myeloma centers and potentially be used for selecting patients into clinical trials.
Paiva:Celgene: Honoraria, Research Funding; Janssen: Honoraria; Takeda: Honoraria, Research Funding; Sanofi: Consultancy, Research Funding; EngMab: Research Funding; Amgen: Honoraria; Binding Site: Research Funding. Mateos:Takeda: Honoraria; Janssen: Honoraria; Amgen: Honoraria; Celgene: Honoraria. Morgan:Univ of AR for Medical Sciences: Employment; Celgene: Consultancy, Honoraria, Research Funding; Janssen: Research Funding; Takeda: Consultancy, Honoraria; Bristol Meyers: Consultancy, Honoraria. Barlogie:Signal Genetics: Patents & Royalties.
Author notes
Asterisk with author names denotes non-ASH members.
This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal