Abstract
Background: There is a global consensus that multiple myeloma patients with high-risk disease require additional monitoring and therapy compared to low/standard risk patients in order to maximize their chances of survival. Current diagnostic guidelines recommend FISH-based assessment of chromosomal aberrations to determine risk status (i.e. t(14;20), t(14;16), t(4;14) and/or Del17p), however, studies show FISH for MM may have a 20-30% QNS rate and is up to 15% discordant between laboratories, even when starting from isolated plasma cells. In this study we demonstrate that MyPRS gene expression profiling reproduces the key high risk translocations for MM risk stratification, in addition to having other significant advantages.
Methods: Reproducibility studies show that MyPRS results are less than 1% discordant starting from isolated plasma cells and return successful results in up to 95% of cases. 270 MM patients from Johns Hopkins University (MD) and Weill Cornell Medicine (NY) had both FISH and MyPRS gene expression profiling performed between 2012 and 2016 using standard and previously published methodology, respectively.
Results: Retrospective review of the matched FISH and MyPRS results showed:
25/28 (89%) patients wish FISH-identified t(4;14) were classified as MMSET (MS) subtype.
10/10 (100%) patients with t(14;16) or t(14;20) were classified as MAF-like (MF) subtype
62/67 (93%) patients with t(11;14) were assigned to the Cyclin D (1 or 2) subtype.
Patients with FISH hyperdiploidy status were classified as the Hyperdiploid (HY) subtype or had multiple gains detected by the separate MyPRS Virtual Karyotype (VK) algorithm, included in MyPRS.
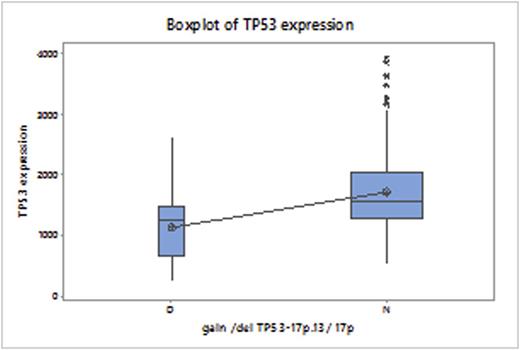
TP53del was seen in patients with multiple molecular subtypes, predominantly Proliferation (PR) and MMSET (MS). Assessment of TP53 function by gene expression is a more clinically relevant prognostic marker than TP53del, as dysregulation of the tumor suppressor is affected by mutations as well as deletions. Analysis of the TP53 expression in the 39 patients with delTP53 showed a statistically significant difference, compared to patients without this deletion (P<0.0001).
Conclusion: Gene expression profiling is a superior and more reliable method for determining an individual patients' prognostic risk status. The molecular subtypes of MM, as reported by Signal Genetics MyPRS assay, are driven by large-scale changes in gene expression caused by or closely associated with chromosomal changes, including translocations.
Physicians who are managing myeloma patients and wishing to base their assessment of risk on R-ISS or mSMART Guidelines may obtain the required data points from either FISH or MyPRS, with the latter offering lower QNS rates, higher reproducibility, assessment of a larger number of cells and a substantially lower price point ($5,480 vs. $1,912; 2016 CMS data). A larger cohort study is now underway to further validate these observations.
GEP-based TP53 expression in patients with and without Del17p. P<0.0001
GEP-based TP53 expression in patients with and without Del17p. P<0.0001
Van Laar:Signal Genetics, Inc.: Employment. Borrelo:Sidney Kimmel Cancer Institute: Employment. Jabalayan:Weill Cornell Medical Center: Employment. Niesvizky:Celgene: Consultancy, Research Funding, Speakers Bureau; Takeda: Consultancy, Research Funding, Speakers Bureau; Onyx: Consultancy, Research Funding, Speakers Bureau. Zielinski:Signal Genetics, Inc.: Employment. Leigh:Signal Genetics, Inc.: Employment. Brown:Signal Genetics, Inc.: Employment. Bender:Signal Genetics, Inc.: Employment.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal