Abstract
Background : Chronic Lymphocytic Leukemia (CLL) is characterized by a very heterogeneous clinical course and the immunoglobulin heavy-chain gene (IgHV) mutational status is currently considered the gold standard of prognostication: unmutated (UM) immunoglobulin heavy chain region (IgHV) is associated with a poor prognosis while patients with mutated IgHV (M) have more indolent disease. An exception are patients with IgHV3-21/IgLV3-21 who have poor prognosis irrespectively of the IgHV mutational status. Interestingly, IgHV3-21 is co-expressed with IgLV3-21 in the majority of cases. However, little is known about IgLV3-21: indeed this light chain has never been characterized independently of IgHV3-21 in terms of gene expression and prognostic impact.
Methods: Based on a cohort of 32 patients with aggressive CLL, we used total RNAseq data of highly purified leukemic cells to define gene expression profiles and IgHV/IgLV rearrangements for each patient. Gene set enrichment analysis was correlated with treatment-free (TFS) and overall (OS) in the initial cohort of 32 patients and in an independent cohort of 255 patients where IgLV3-21 positivity was determined by real-time PCR and confirmed by Sanger sequencing.
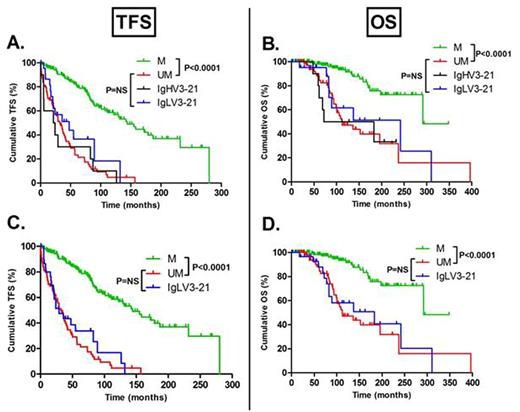
Results: Among the 32 initial CLL patients, 9 patients had an IgLV3-21 rearrangement, but only 1 patient carried the IgHV3-21 rearrangement. The other patients had VH1-24/69, VH3-9/66/23/48/53 and VH4-59 heavy chain rearrangements. RNAseq expression profiling yielded 1457 transcripts and 789 genes that were differentially expressed between IgLV3-21 patients and the other patients. Within the differentially expressed genes, 68% were upregulated while 32% were downregulated at least 1.5 fold. Gene set analysis revealed enrichment of genes related to translation enhancement (ribosome, translational reactome, peptide chain elongation, RNA metabolism - P<0.0001) and MYC target genes (P=0.0003), in line with recent finding showing that BCR stimulation can increase global mRNA translation including MYC-specific mRNA translation. In the initial cohort of 32 patients, IgLV3-21 patients had a median TFS of 17 months compared to 44 months in patients with another light chain (P=0.0270). Similarly, IgLV3-21 patients had a shorter median OS (88 months vs >192 months, P=0.0287).We validated these results in an independent cohort of 255 patients with 31 (12%) IgLV3-21 patients and 10 (4%) with IgHV3-21 (of which 8/10 also carried the light chain IgLV3-21 rearrangement). IgLV3-21 patients presented a median TFS/OS of 29/183 months compared to non IgLV3-21 patients who had a median TFS/OS of 88/292 months (P=0.0003/P=0.0142). In addition, when IgHV3-21 patients (n=10) were compared to IgLV3-21 only patients (n=23), no statistical difference was observed in terms of TFS or OS. Interestingly, if patients were classified according the IgHV mutational status, both IgHV3-21 and IgLV3-21 patients displayed a prognosis similar to UM patients: median TFS was 144, 32, 23, 48 months for M, UM, IgHV3-21 and IgLV3-21 patients, respectively (Figure A- P<0.0001). Similar results were observed for OS with a median OS of 292, 112, 128 and 241 months for M, UM, IgHV3-21 and IgLV3-21 patients, respectively (Figure B - P<0.0001). If all IgLV3-21 (n=31) were considered independently of their heavy chain, median TFS (29 months) were similar to UM patients (32 months, P=0.5536) and statistically different from M patients (144 months - P<0.0001, Figure C). Similar results were observed for OS (Figure D).
Conclusions: Our results highlight for the first time the importance of light chain IgLV3-21 in CLL in terms of its differential gene expression profile and prognosis: IgLV3-21 is associated with a translational enhancement gene signature and confers a poor prognosis similar to UM patient irrespectively of the heavy chain IgHV3-21 or the IgHV mutational status.
Schuh:Gilead: Consultancy, Honoraria, Research Funding; Roche, Janssen, Novartis, Celgene, Abbvie: Consultancy, Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal