Abstract
Background: Survival of pts with chronic lymphocytic leukemia (CLL) ranges from a few years to a normal lifespan. Clinical stages (i.e. Rai, Binet) are the basis for CLL prognostication but do not reflect the complex biology of CLL, which ultimately shapes the disease heterogeneity. Recently, a CLL-International Prognostic Index (CLL-IPI) which includes five clinical and biological variables (i.e. age, IGHV mutational status, del(17p), beta2-microglobulin [B2M] and Rai or Binet stages) and stratifies pts into four different categories has been proposed. This prognostic index is obtained by the sum of the score given to each parameter and includes dichotomized continuous variables. The aim of this study was to determine whether a prognostic model based only on biomarkers could separate CLL patients with different outcomes and simplify the CLL-IPI.
Material and Methods: Five hundred twenty-four CLL pts from the Hospital Clínic, Barcelona, in whom information at diagnosis included age, clinical stage (Rai and Binet), IGHV mutational status, B2M, and FISH-cytogenetics were analyzed. For validation purposes a cohort of 417 pts from the Brno Hospital, Czech Republic was used. The two series included patients as seen in clinical practice. Primary endpoints were overall survival (OS) and time to first treatment (TTFT). The internal validity of the models was evaluated using bootstrapping, and the discriminatory value by c-statistics. Double sided P values < .05 were considered significant.
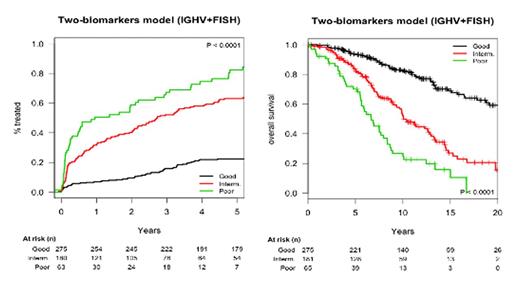
Results: First, we confirmed that all five covariates included in the CLL-IPI were independently predictive of OS. Both sets of five covariates (age + B2M + IGHV + FISH + Rai or Binet) were incorporated into the regression models. All four patient subgroups had a significantly different survival (c-statistic: 0.72), although the high- and very-high risk groups overlapped and the number of patients in the very-high-risk group was small. We then evaluated all possible combinations of these five covariates to identify the simplest model with robust discriminatory value. A prognostic model based on IGHV mutational status + FISH [del(17p) and/or del(11q)] separated three subgroups of pts [i.e, good-biomarkers (mutated IGHV + no poor FISH cytogenetics), intermediate-biomarkers (either unmutated IGHV or poor FISH cytogenetics), and poor biomarkers (unmutated IGHV + poor FISH cytogenetics)] with different prognosis (c-statistic: 0.68). In the Barcelona series, the good-biomarkers category identified around 50% of pts from the whole series whose survival did not differ from the general population; patients with intermediate-biomarkers had a projected 10-year survival of 68%. Finally, the poor-biomarkers category captured pts with a projected 10-year survival of 17%. The corresponding TTFTs at 3 years from diagnosis were 16%, 50% and 63%, respectively (Figure). Furthermore, it separated patients with different outcome within clinical stages, notably Binet A or Rai 0, and across all age groups. This model was fully validated in the Brno series.
Conclusions: The biomarkers-only CLL prognostic model presented here is based on the two most important CLL biomarkers (i.e. IGHV mutational status and FISH cytogenetics) which are included as a backbone in the CLL-IPI and other CLL prognostic systems. This model is simple, easy to apply and to remember; it separates three groups of pts rather than four; it does not contain continuous variables; it can be applied to younger and older pts, and has clinical implications. This prognostic model could be useful in CLL prognostication either alone or in combination with clinical stages and warrants prospective validation, including in patients receiving targeted therapies.
Montserrat:Pharmacyclics: Consultancy; Vivia Biotech: Equity Ownership; Janssen: Honoraria, Other: travel, accommodations, expenses; Gilead: Consultancy, Other: Expert Testimony; Morphosys: Other: Expert Testimony.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal