Abstract
Background: The recent advances in molecular techniques and the adaptation of next generation sequencing (NGS) in routine clinical testing increased our ability to use molecular approaches in the diagnosis and classification of most hematologic diseases. Bone marrow aspiration and biopsy remains necessary for initial confirmation of diagnosis of neoplastic processes in bone marrow, but significant literature suggests that screening or monitoring patients by testing peripheral bloodcfDNAmight be a reliable alternative to marrow biopsy and might reduce the need for a painful bone marrow procedure. Here we report the results of routine clinical testing ofcfDNAthat is ordered by practicing hematologist in the context of the presence or the suspicion of the presence of hematologic neoplasm.
Methods: A total of 227 peripheral blood samples were submitted for screeningcfDNAfor mutations in a 54 gene focusedMyeloidpanel using NGS sequencing. DNA was extracted from plasma usingNucliSenSEasyMAGautomated platform and then assayed using theTruSightMyeloid Sequencing Panel (Illumina; San Diego, CA) with an average sequencing depth of 10,000X. The average age patients was 71 (18-96) years. The reason for submitting samples wasruling out MDS in 199 and ruling out AML or other hematologic neoplasms in 28 samples. Of these samples, 12 patients had a follow up testing of bone marrow aspiration sample.
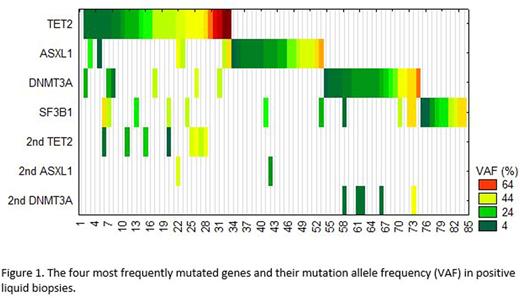
Results: Of the 227 tested samples (Figure 1), 126 (55%) showed no evidence of mutation in any of the tested genes. Based on our previous data (see ASH abstract by Albitar et al, 2016), this suggested that MDS can be ruled out in these patients and bone marrow biopsy could be avoided and not recommended. In contrast, 101 (45%) had mutations in one or more genes. Twenty-nine (~12.8%) contained a mutation in a single gene with variant allele frequency (VAF) <20% in one gene and were considered not diagnostic for the presence of clinically significant hematologic neoplasm, but follow up was recommended. Of these patients, one had a mutation in JAK2 at VAF of 6% and a second had a mutation in CALR gene at VAF of 7%, which most likely suggest the presence of early evolvingmyeloproliferativeneoplasms. Seventy-four patients (33%) had mutations in two or more genes or in one gene but with VAF≥20% and considered diagnostic for the presence of hematologic neoplasm and bone marrow morphologic evaluation was recommended. The most commonly mutated gene in these patents was TET2, detected in 30 samples, of which 8 also showed a second mutation in TET2, followed by ASXL1, and DNMT3A mutated in 24 and 26 samples, respectively. Samples containing a TET2 mutation were more likely to have a second mutation in TET2 or another gene, in contrast other genes that were frequently mutated did not show this trend (see Figure). TP53 gene was mutated in 16 samples, 7 of which as a single abnormality with VAF <20% therefore was reported as of unknown significance and recommended ruling out neoplasms in hematologic (lymphoid and myeloid) as well as solid tumors. SF3B1 gene mutations were detected in 19 samples and recommended ruling out refractory anemia with ringsideroblasts(RARS). Despite the small sampling (12 samples), follow up usingcfDNAtesting reliably recapitulated original bone marrow Biopsy's findings. In one patient, additionalsubcloneswere detected incfDNAthat were not detected in the bone marrow aspirate.
Conclusions:cfDNAtesting is reliable approach to screen for the presence of Hematologic neoplasm and potentially could avoid the need for bone marrow biopsy in almost half the patients expected to have MDS or otherhematopoeticneoplasms. Positive diagnosis can be confirmed in additional 45% of patients and only 12.8% of patients will be reported with questionable results. Except for those with TP53 mutations, the rest of the 12.8% cases can be classified as Clonal Hematopoiesis of Indeterminate Potential (CHIP). While bone marrow is still the gold standard, our real world experience shows liquid biopsies can be sensitive and non-invasive approach to rule out MDS or other hematological diseases.
Funari:NeoGenomics Laboratories: Employment. Ma:Neogenomics Laboratories: Employment. Thangavelu:Neogenomics Laboratories: Employment. De Dios:Neogenomics Laboratories: Employment. Albitar:Neogenomics Laboratories: Employment, Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal