Abstract
Primary cold agglutinin disease (CAD) is a hemolytic anemia mediated by monoclonal anti-I autoantibodies. CAD is caused by an underlying low grade B-cell lymphoproliferative disease of the bone marrow with a typical histology that is different from lymphoplasmacytic lymphoma and, accordingly, does not display the MYD88 L265P mutation (Randen et al., Haematologica, 2014).
Since CAD is a clonal lymphoproliferative disorder, we studied the mutational landscape to further characterize the disease and identify potential novel treatment approaches.
We prospectively collected bone marrow samples of CAD patients, enrolled in a clinical trial (CAD5; www.clinicaltrials.gov, NCT02689986). Exome sequencing of six cases was performed and findings were confirmed in ten additional cases using targeted sequencing. For these analyses, clonal B cells and normal T cells, used as control, were purified from bone marrow samples using fluorescent activated cell sorting. All mutations were verified by Sanger sequencing. Whole-exome sequencing was performed at BGI Tech Solutions (Hongkong) using the Agilent SureSelect Human All Exon V4 Reagent Kit and Illumina HiSeq technology. The bioinformatics pipeline consistent of BWA alignment tool (aligned to hg19); Picard tools FixMateInformation and MarkDuplicates; the Genome Analysis Toolkit (GATK) IndelRealigner and BaseRecalibrator; somatic variant detection tools Strelka, MuTect and Pindel; the annotation tool SnpEff.
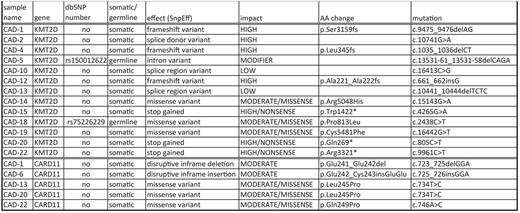
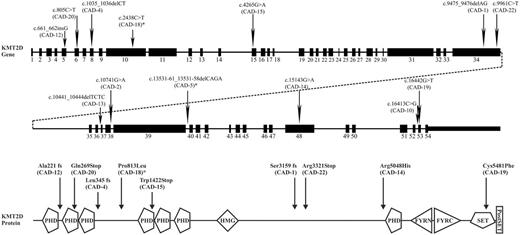
Recurrent somatic mutations were found in KMT2D (11/16 cases, 69%) and CARD11 (5/16, 31%) (Table 1, Figure 1-2). 7/16 (44%) of KMT2D mutations were deemed high impact mutations by SnpEff and to result in inactive protein. 2/16 (12,5%) of KMT2D mutations are missense mutations and are predicted to impair SET domain function. 2/16 (12,5%) of KMT2D mutations are classified by SnpEff as low impact mutations of which functional tests are necessary to demonstrate potential consequences for protein function. Of interest, two additional patients showed rare germline KMT2D variants that have also been seen in Kabuki syndrome patients, although these patients do not have Kabuki syndrome.
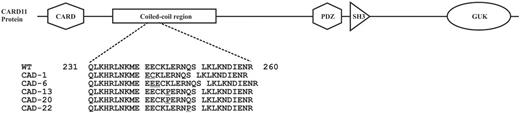
CARD11 was somatically mutated in 5/16 (31%) cases. Four of those patients had a concurrent KMT2D mutation. All CARD 11 mutations were classified as moderate impact mutations by SnpEff. Mutations were tightly clustered in a 20bp sequence of the coiled-coil domain sequence (Table 1, Figure 2).
KMT2D is a histone lysine methyl transferase that represses B cell lymphoma development. Mono-allelic mutations of KMT2D seem to act in a dominant fashion and cause partial loss of protein expression with cell growth advantage. KMT2D is frequently mutated in follicular lymphoma, diffuse large B cell lymphoma and nodal marginal zone lymphoma. Mono-allelic constitutional mutations cause Kabuki syndrome, characterized by distinct facial characteristics and multiple organ malformations. Patients frequently develop immune-mediated thrombocytopenia as well as auto-immune hemolytic anemia.
CARD11 coiled-coil domain mutations result in constitutive NF-kB activation and enhanced NF-kB activity upon antigen receptor stimulation. Mutations were previously detected in diffuse large B cell lymphomas of activated B cell origin. Mono-allelic CARD11 coiled coil mutations are not oncogenic per se in mice and humans, but result in B-cell proliferation and auto-antibody production. Since four CAD patients showed concurrent KMT2D and CARD11 mutations, it seems likely that the mutations act in concert with anti-I B-cell receptor stimulation to contribute to CAD-associated lymphoproliferative disease.
In conclusion, we demonstrated a high frequency of KMT2D and CARD11 mutations in the bone marrow B cell lymphoproliferative disease of patients with CAD. These results confirm that CAD-associated B cell lymphoproliferative disease is a distinct disease different from other known B cell lymphoproliferative diseases of the bone marrow, most notably lymphoplasmacytic lymphoma. The identification of these recurrent mutations in CAD may allow the design of novel treatment modalities.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal