Abstract
Background: The PI3K pathway plays a significant role in cell cycle, apoptosis and DNA repair and it is commonly dysregulated in cancers making it an attractive therapeutic target. BKM120 (buparlisib) is a novel oral pan-class I PI3K inhibitor with antitumor activity and efficacy reported in solid tumors. It is in phase I/II clinical trials for treatment of relapsed-refractory NHL. There is a paucity of data, however, on molecular mechanisms of action and biological pathways of resistance for BKM120-treated lymphoma. Through a systems biology approach, we determined key molecular pathways and mechanisms associated with BKM120 treatment in a multitude of lymphoma cell lines.
Methods: T cell lymphoma (Jurkat, Hut78, HH), HL (L428, L540), Burkitt lymphoma (Raji), and DLBCL (SUDHL4, SUDHL10, OCILY19) cell lines were treated with increasing concentrations of BKM120 (0.16-10uM) in 96 well plate and cell viability assessed by MTT assay. Global transcriptome analysis was done on SUDHL4, Raji, Jurkat, Hut78 and L540 cells treated with BKM120 at IC50 and compared to untreated control cells. Significant genes were determined by applying a one-way ANOVA with an adjusted Bonferroni correction for a false discovery rate (FDR) < 0.05 with further analysis using a fold change greater than ±1.2. Pathway and functional relationships were observed using Ingenuity Pathway Analysis and Gene Set Enrichment Analysis. Established systems biology analysis technique was used to determine key genes involved with BKM120 treatment. Apoptosis was evaluated by Annexin V and propidium iodine (AV/PI) analysis. For cell cycle analysis, cells were exposed to BrdU and stained with anti-BrdU FITC and 7-AAD and analyzed by flow cytometry.
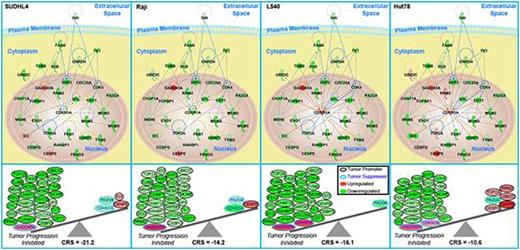
Results: Cell viability showed dose-dependent growth inhibition in all lines with IC50 between 0.316μM-3.72μM at 72 hours. Global transcriptome analysis revealed BKM120 treated lymphoma cells had a conserved inhibitory impact on cell cycle, DNA replication and metabolic process across all cell lines. Systems biology analysis revealed 32 key driving genes responsible for BKM120 predicted inhibition of tumors for all lymphoma cells with exception of Jurkat (see figure below). GADD45A, a known tumor suppressor involved with DNA damage response, was found as a key upregulated gene involved in BKM120-treated cells. Although a predicted inhibition for tumor growth was also predicted for Jurkat, a different profile of key genes was seen. AV/PI-staining and flow cytometry revealed dose-dependent increase of apoptotic cells in all lymphoma cell lines. BrdU incorporation revealed induction of G2/M arrest in Jurkat and Hut78. GADD45 siRNA knock out in Hut78 cells treated with BKM120 abrogated G2/M arrest. Western blots showed decreased phosphorylation of PI3K and mTOR substrates including 4-EBP and phospho-p70S6K with BKM120 treatment. Phosphorylation of MEK/ERK was down regulated at lower doses of BKM120, while increased phosphorylation was noted at higher doses. Finally, analysis of cell cycle regulatory proteins Cyclin, CDK and phospho-histone H3 with BKM120 treatment resulted in expression changes consistent with G2/M arrest and led to increased PARP cleavage in all lymphoma cell lines.
Conclusions: Through systems biology analyses, we identified that BKM120 treatment of lymphoma produced prominent impairment of cell cycle, DNA replication and metabolic function as core responses to PI3K inhibition, which contributed to tumor inhibition predictions. Possible mechanisms of resistance to BKM120 were predicted through cell cycle (G2/M) arrest and MAPK activation. Furthermore, GADD45A was discovered as a key driver with BKM120 treatment across multiple lymphoma cell lines. Collectively, these data provide strong pre-clinical evidence towards identification of key drivers responsible for resistance with PI3 kinase inhibitor therapy, which may inform biomarker discovery as well as rational therapeutic combinations.
Common key genes for BKM120 treated. Top panels are networks of the key genes interacting. Bottom panels are the prediction for cancer risk with BKM120 treatment versus controls. Based on literature the fold-change values of genes are classified as tumor promoters or tumor suppressors. Carcinogenic Risk Score (CRS) is calculated based on these values to determine if there is a promoted risk (positive value) or inhibitory risk for cancer (negative value).
Evens:Takeda: Other: Advisory board.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal