Abstract
Background. We have recently investigated transcriptional and posttranscriptional changes in AML cells (Oncotarget, 2016, 7, 2889-909) and identified ABCA3 expression, TET2 expression and TET2 exon 2 skipping (TET2E2S) as prognostic factors in pediatric AML (See companion abstracts from Ceraulo et al.). The present study was conducted in order to test whether the combination of these prognostic factors might help stratifying patients according to their relapse risk.
Methods. Samples derived from 120 patients with available high-quality RNA and enrolled in the French ELAM2 protocol. Patients were classified according to their standardized cytogenetic and molecular (NPM1 mutations, FLT3-ITD, CEBPA double mutations) risk subgroups. Treatment consisted of 1 induction course (AraC and mitoxantrone) and 3 consolidation courses (course 1 and 3 with high dose AraC); all children with either intermediate or high-risk disease were candidates for hematopoietic stem cell transplant in complete remission (CR) after 1 to 2 consolidation courses. qRTPCR amplification of 2 conserved ABCA3 (exons 6-7 and exon 19-20) and TET2 (exon 4 and exon 2-3) mRNA sequences were performed with GUS and ABL as reference genes. TET2E2S was quantified as already described (Oncotarget, 2016, 7, 2889-909). The concordance index (c-index) was used to compare the relative statistical power of the new model compared with the standardized cytogenetic and molecular-based scoring system.
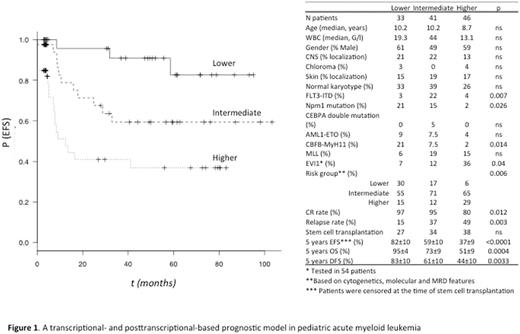
Results. ABCA3 expression [HR 2.6 (95% CI 1.3-5), p=0.006], TET2 expression (HR 0.5 (0.2-0.9) p=0.04, and TET2E2S (HR 2.5 (1.2-5.1), p=0.01) represented independent prognostic factors for EFS. To design a convenient prognostic score, continuous variables were dichotomized using cutoff points that were based using the Maximally Selected Rank Statistics (maxstat R-statistics package, from the R-Project). One point was attributed for lower TET2 expression (n=85), higher ABCA3 expression (n=44), and TET2E2S (n=61). Three groups were formed: lower-risk patients (0 point, n=33), intermediate-risk patients (1 point, n=41), and higher-risk patients (either 2 or 3 points, n=46). The present score identified subgroups of patients with significantly different outcome (p<10-4) (Figure 1). Patients with a low TPS (Transcriptional-Posttranscriptional Score) had significantly better EFS than either patients with a high (p<10-4) or an intermediate TPS (p=0.03) while patients with intermediate TPS had significantly better EFS than patients with high TPS (p=0.02). For OS (p=0.0004), patients with a low TPS had significantly better OS than either patients with a high (p=0.0004) or an intermediate TPS (p=0.047) while patients with intermediate TPS had significantly better OS than patients with high TPS (p=0.024). For DFS (p=0.003), patients with a low TPS had significantly better DFS than either patients with a high (p=0.0012) or an intermediate TPS (p=0.036) whereas no significant difference was noticed between patients with intermediate versus high TPS (p=0.2). Figure 1 shows that FLT3-ITD was significantly associated with intermediate TPS, EVI1 expression with high TPS and NPM1 mutations and CBFB-MyH11 leukemias with low TPS. The distribution of CR rate and relapse rate was significantly distinct among the 3 TPS categories (Figure 1). The added value of the TPS within AML genetic risk groups was evaluated through univariate analysis. The TPS permitted to identify patients with different outcome in lower and intermediate-risk AML (EFS: Log-rank, p=0.011 and 0.005, respectively). Ten of the 20 lower-risk AML corresponded to intermediate (n=7) and high (n=3) TPS. Their CR rate was 100%, none of these patients was allografted and 4 patients relapsed as compared to 0/10 in the remaining lower-risk AML cases with low TPS (p=0.02). In intermediate risk AML, the CR and relapse rates of patients with low, intermediate and high TPS were 94 and 17%, 100 and 45%, and 73 and 50 %, (CR, p=0.001; relapse, p=0.036). Overall, the present TPS model had a better c-index (0.73) compared with the standardized cytogenetic and molecular-based scoring system (0.61).
Conclusion. We propose a new prognostic model that includes transcriptional and posttranscriptional data and can predict survival in pediatric AML. A validation in a larger, independent, multicenter, data set of patients is important to facilitate the translation of this model into clinical practice.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal