Abstract
Background: Acute myeloid leukemia (AML) is initiated and maintained by a subpopulation of leukemic stem cells (LSCs), which are defined by their property to serially engraft immunocompromised mice. Based on this a LSC-associated gene expression was identified which was shown to be prognostically relevant in a retrospective analysis (Eppert et al, Nature Medicine 2011). microRNAs (miRs) play important roles in leukemogenesis, serve as potential tumor markers and novel therapeutic targets. In the current analysis we wanted to establish a miRNA profile of functionally validated LSCs from AML patients of various molecular and cytogenetic subgroups by conducting a comprehensive functional genomics study.
Method: In total, 17 de novo AML samples were sorted for CD34+ granulocyte-macrophage progenitor (GMP), CD34+ lymphoid-primed multi-potential progenitor (LMPP) and CD34- subpopulations. Sorted cells were transplanted into sublethally irradiated non-obese diabetic/severe combined immunodeficient Gamma (NSG) mice. Those subpopulations that gave rise to human leukemic cell engraftment > 1% were classified as LSCs and sub-divided into LMPP-LSCs and GMP-LSCs and compared to the CD34- cell compartment which did not give rise to human leukemic cell engraftment (non-LSC). In parallel corresponding counterparts were highly purified from healthy bone marrow (BM). miRNA libraries were prepared using the Illumina TruSeqTM Small RNA Sample Preparation Kit and sequenced as single end reads on a Illumina HiSeq 2000. The bioconductor package limma was used for identifying differentially expressed miRNAs, the made4 package for correspondence analysis and between group comparisons. Quantitative RT-PCR validation on bulk and sorted populations (LMPP-LSC, GMP-LSC, CD34-) tested the expression levels of the identified miRNAs.
Results: We assessed the engraftment potential of 17 AML samples, sorted into the LMPP, GMP and CD34- fraction. The LMPP or GMP fractions of 8 patients showed a leukemic graft in the NSG mouse model, whereas the CD34- fractions were not able to show a human engraftment in these mice. To the end subpopulations of 8 validated patient samples and corresponding stem-, progenitor, and CD34- subpopulations of normal bone marrow samples (n=3) were analyzed. Global miRNA expression profiles of subpopulations from normal and patient samples were compared using correspondence analysis based between group analysis. First, LSC subpopulations were compared to the leukemic CD34- Non-LSCs: between the LMPP-LSCs and the CD34- non-LSCs 15 miRNAs were differentially expressed in the LMPP-LSCs; between the GMP-LSCs and the CD34- non-LSCs only six miRNAs were found to be differentially expressed. Using these miRNAs, an unsupervised hierarchical clustering showed that the LMPP-LSCs, the GMP-LSCs and leukemic CD34- samples formed non-overlapping clusters. In normal bone marrow, 12 miRs showed differential expression between normal BM LMPP and CD34- population and 36 miRNAs between BM GMP vs. BM CD34- cells.
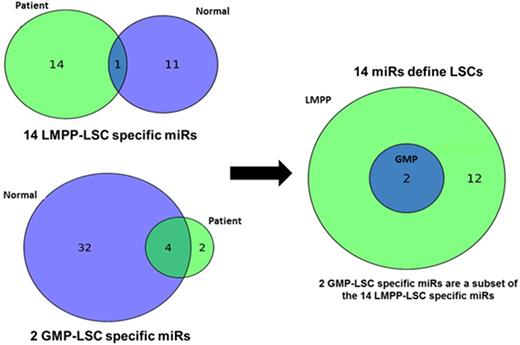
We then compared the miRs that were differentially expressed in the patients compared to the differentially expressed miRs between the normal BM populations: except for one miR (miR-182), the fifteen differentially expressed miRs in the LMPP-LSC versus leukemic CD34- samples did not show overlap with the twelve miR significantly differentially expressed between normal BM LMPP and CD34- cells. When the GMP-LSC to leukemic CD34- Non-LSC comparison was cross-compared to the BM GMP to normal BM CD34- comparison, only two miRs (miR-148a, miR-320a) were unique to the patient samples. When we finally compared the LMPP-LSC specific 14 miR to the 2 GMP-LSC specific miRs, interestingly, we observed that the GMP-LSC miRs were a subset of the LMPP-miRs. Thus, 14 miRs defined a set of miRs differentially expressed in functionally validated AML LSCs. Importantly, 4 of these 14 miRs represent miR-941 isoforms. This miR has not been linked to leukemogenesis yet. It is one of the very few human-specific miRs which developed de novo in the human lineage after separation of the human and the chimpanzee lineages between six and one million years ago and preferentially targets genes in hedgehog- and insulin-signalling pathways. Taken together, combining functional validation of AML LSCs with next generation sequencing allows to identify miRs so far not linked to AML biology and LSC properties.
Mulaw:NuGEN: Honoraria. Buske:Celltrion, Inc.: Consultancy, Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal