Abstract
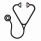
Gemtuzumab ozogamicin (GO), a CD33-targeted immunoconjugate, is a re-emerging as therapeutic for AML. We have previously discovered polymorphisms in CD33 coding region that might be associated with outcome in patients treated with GO. One particular coding polymorphism, CD33-SNP rs12459419-C>T (Ala14Val), is located within the splice enhancer region of exon-2, leading to expression of an alternate splice isoform lacking exon-2. This alternate splice isoform (D2-CD33), would encode a protein product lacking the IgV domain, which is the binding site for GO and most if not all CD33 antibodies used for diagnostic immunophenotyping. We therefore hypothesized that the SNP rs12459419 genotype would be associated with differential expression of the D2-CD33 transcript levels and differential cell surface CD33 expression leading to genotype determined differential response to GO.
We evaluated the genotype frequency and functional significance of rs12459419, its association with CD33 cell-surface expression on leukemic blasts, and clinical response in 816 children and young adults with AML randomized to GO vs. No-GO in the COG trial AAML0531.
The CD33 SNP rs12459419 genotype frequency was-CC=51%, CT=39% and TT=10% in patients, similar to the observed frequency in the general population. Correlation of SNP allele frequency with CD33 transcript levels and surface CD33 expression (as determined by p67.6 antibody) demonstrated that the T-allele was significantly associated with higher levels of D2-CD33 transcript (P=4.7e-11, Figure 1) and with lower diagnostic leukemic cell CD33 surface intensity (P=1.93e-29).
Clinical outcome based on the SNP genotype demonstrated that patients with CC-genotype had significantly lower RR of 26%±7% in the GO arm whereas those in the No-GO arm had a RR of 49%±9% (HR=0.468, P<0.001). The corresponding DFS for CC genotype patients in the GO and No-GO arms was 65%±7% and 46%±9%, respectively (HR=0.597, P=0.004; Figure 1). In contrast, in those with heterozygous CT or homozygous TT genotype, GO exposure provided no clinical benefit in RR (CT: 38%±9% vs. 37±10%, P=0.975; TT: 46%±20% vs. 46%±20%, P=0.798) nor DFS (CT: 56%±9% vs. 60%±10% GO vs. No-GO, P=0.821; TT: 51%±20% vs. 54%±18%, GO vs. No-GO, P=0.972, Figure 1).
We further evaluated the impact of the CD33 genotype on the efficacy of GO in different risk groups as well as in high vs. low CD33 expression cohorts. Patients in the low-risk (LR) group with the CC genotype treated with GO had a RR of 10%±8% vs. 37%±13% (P< 0.001) from remission. Standard-risk patients with CC genotype had a RR of 41±12% vs. 59±12% (P=0.056) and high-risk patients had a RR of 36%±27% vs. 70%±32% (P=0.073) for the GO and No-GO arms. In contrast there was no benefit of addition of GO in patients with the CT or TT genotypes within each risk group.
Since CD33 expression has been recently associated with GO efficacy, we evaluated the association of rs12459419 genotypes in patients with low (N=153) and high CD33 (N=436) cell surface expression quartile-1 and quartiles-2-4, respectively. Significant improvement in RR was observed in patients with the CC genotype in the GO arm over those in the No-GO arm (P=0.001) within quartiles 2-4, but not with the CT/TT genotype (P=0.112). Within lower CD33 expression (quartile-1) cohort, a similar trend towards improvement in RR compared with those in the No-GO arm (P=0.055) was observed although the CC genotype was less frequent.
Multivariate cox regression analysis that included genotype, risk status, and CD33 expression demonstrated that CD33 CC genotype was an independent predictor of response to GO (HR= 0.45, P<0.001 for RR and HR=0.57, P=0.003 for DFS).
The rs12459419 genotype mediates expression of the GO binding site and informs on which patients should receive GO. The knowledge of CD33 genotype and prediction of response to GO provides opportunities to use patient genotypes for selecting CD33-targeted therapies. Given only half of the patients are expected to have a response to GO, we propose that all prior GO containing studies be re-evaluated for response based on patient CD33 genotype. The efficacy of GO in patients with an appropriate antibody binding domain also raises the possibility of developing next-generation CD33 immunoconjugates with epitopes targeted to regions not affected by alternative splicing and SNPs.
Loken:Hematologics: Employment, Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.
This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal