Abstract
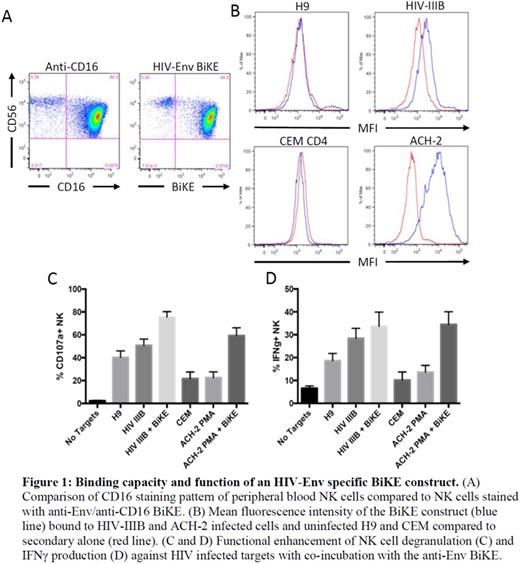
Natural Killer (NK) cells, a critical component of the immune response to viral infection, recognize and destroy cells with diminished expression of major histocompatibility class-I (MHC-I) molecules and expression of ligands for activating NK receptors such as NKG2D. Down-modulation of MHC-I is a hallmark of viral infection, as it allows infected cells to evade a CD8 T-cell response. Stalling of the cell cycle to enhance viral replication induces NK activation ligands such as the NKG2D ligands unique long binding proteins (ULBP)-1 and -2 which could trigger NK destruction of infected cells. Unfortunately, incomplete down-modulation of MHC-I by HIV leaves HLA-C on the cell surface, which inhibits the majority of NK cells from killing infected targets. CD16, the low affinity Fc receptor, is the most potent NK cell activating receptor. It mediates antibody dependent cell-mediated cytotoxicity (ADCC), and can override inhibition by MHC-I. We designed a series of bi-specific killer-engager (BiKE) constructs to direct NK cell ADCC against an HIV-infected target. We linked the Fab portions of broadly neutralizing (bn)Abs to a novel llama-derived nanobody EF91 that binds CD16 at high affinity and signals strong activation. We chose to use EF91 as its structure is unique compared to the use of a single chain variable fragment (scFv). Rather than being composed of a variable heavy (VH) and variable light (VL) chain, the nanobody is composed of a single variable heavy (VHH) domain. A distinct advantage to using a CD16 nanobody over a scFv is in the purity of the generated product. During protein folding it is not uncommon for the wrong VH to associate with the wrong VL; the result of which is a nonfunctional product. Since the nanobody is single VHH, and does not require association with another domain, there is less risk of a misfolded product. Nanobodies are also known to have similar, if not increased, affinity for their target molecules. In the case of EF91, this may result in more robust activation of NK cells than with a traditional scFv. We tested a BiKE constructed with the bnAb, VRC01, which recognizes the CD4 binding domain of HIV-Env. The specificity of our novel anti-CD16 nanobody was demonstrated by binding of our BiKE construct to CD16+ NK cells (Figure 1A). Function of our BiKE construct was tested by incubating it with chronically infected T-cell lines (HIV-IIIB and ACH-2) or with their respective uninfected counterparts (H9 and CEM). We only observed binding to infected cells (Figure 1B), demonstrating HIV-Env binding specificity to the HIV strains ACH-2 (LAI strain) and HIV-IIIB. The ability of the anti-Env BiKE construct to mediate ADCC and IFNγ production was tested against two uninfected CD4 T-cell lines or their infected counterparts. While NK cells degranulated when incubated with the infected cell lines (50% against HIV-IIIB and 20% against LAI), this response was markedly enhanced when co-incubated with the HIV-Env specific BiKE (80% against HIV-IIIB and 60% against LAI) (Figure 1C). Furthermore, the HIV-Env BiKE enhanced IFNγ production against HIV-infected T-cell lines compared to responses in the absence of BiKE (28% against HIV-IIIB compared to 36% with BiKE; 15% against ACH-2 compared to 37% with BiKE) (Figure 1D). Our data demonstrate that a BiKE construct containing the Fab of an HIV bnAb and an anti-CD16 component can eliminate HIV-infected targets that express the HIV-envelope on their surface. The reservoir of latently infected CD4 T cells lack expression of any recognizable virus protein on the cell surface, we plan to combine our BiKE strategy with cellular activation using IL-15. Alternatively, we can construct a tri-specific engager (TriKE) with an IL-15 segment that may activate CD4 T cells while enhancing NK cell killing.
Cooley:Fate Therapeutics: Research Funding. Vallera:Oxis Biotech: Consultancy, Membership on an entity's Board of Directors or advisory committees. Miller:Fate Therapeutics: Consultancy, Research Funding; Oxis Biotech: Consultancy, Other: SAB.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal