Abstract
Introduction: Multiple myeloma (MM) is the second most common hematologic malignancy. Recent sequencing studies show evidence of massive genetic heterogeneity reflected in multiple parallel subclones already at diagnosis. Different subclones respond differently to given therapy. The role of treatment-driven subclonal skewing and intrinsic acquired mutations is poorly understood in the relapse setting. At relapse, the distribution of subclones throughout the bone marrow and at extramedullary sites is currently unknown. Methods: We performed a research autopsy on a single individual with IgG kappa MM who survived with multiple metastatic sites of disease for 10 years after diagnosis. Genomic DNA was extracted from histologically confirmed snap frozen normal tissue and nine independent sites of extraosseous disease. Whole exome sequencing (150X) was performed by the MSK Genomics Core. Data were filtered to remove germline polymorphisms and enrich for high quality somatic variants. Phylogenetics and subclonal analyses were performed using Treeomics and SCHISM.
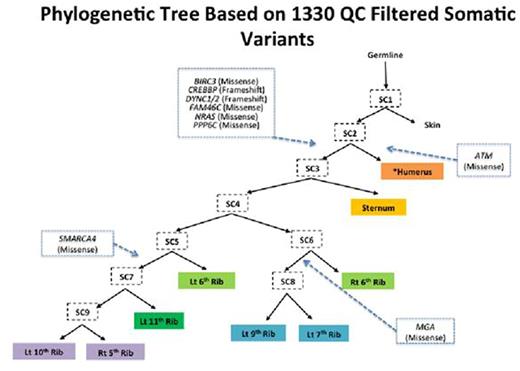
Results: The average on target coverage was 177X with 73% of bases covered a minimum of 100x. A total of 6348 somatic variants were initially identified. After filtering for high quality somatic variants, 1330 remained with 220 of these variants common to all MM samples and an average of 628 variants per sample. Annotation of high quality variants revealed potentially deleterious somatic mutations in NRAS, FAM46C, DYNC1, CREBBP, ATM, BIRC3, MGA, PPP6C and SMARCA4. Phylogenetics analyses (shown below) indicated the metastases arose through a combination of branched and parallel evolution, with the ATM mutation arising in one branch (one metastasis) that was distinct from a second clonal population containing the NRAS, FAM46C, BIRC3, CREBBP, DYNC1 and PPP6C mutations that were present in all eight other metastases. The MGA and SMARCA4 mutations further identified two additional subclones arising in from the NRAS/FAM46C mutant common ancestral clone. Clonality analyses independently supported this hierarchy by identifying NRAS and FAM46C as early clonal events and MGA and SMARCA4 as late events in the progression of this patient's disease. It also suggested a degree of subclonal mixing within each metastatic site.
Conclusions: Using whole exome sequencing from nine independent sites of extraosseous disease in a single MM patient with relapse 10 years after initial diagnosis, we show that extramedullary disease arise through a combination of branched and parallel evolution. Two additional patients have also undergone research autopsy and results will be presented at the meeting.
Landau:Prothena: Honoraria, Membership on an entity's Board of Directors or advisory committees; Takeda: Membership on an entity's Board of Directors or advisory committees, Research Funding; Onyx/Amgen: Research Funding; Spectrum Pharmaceuticals: Honoraria, Membership on an entity's Board of Directors or advisory committees; Janssen: Consultancy. Hassoun:Binding Site: Research Funding; Novartis: Consultancy; Celgene: Research Funding; Takeda: Consultancy, Research Funding. Korde:Medscape: Honoraria. Landgren:Medscape Myeloma Program: Honoraria; Takeda: Honoraria; Amgen: Honoraria, Research Funding; BMS: Honoraria; Merck: Honoraria; Celgene: Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal