Abstract
Chronic lymphocytic leukemia (CLL) is the most common blood cancer in adults and known for its heterogeneous disease course. The nature of CLL calls for new studies of pathogenetic variables validated by prognostic impact. Here we propose a new biological classification system of CLL that associates normal B cell subsets gene expression (GE) signatures (BAGS) with GE in CLL.
We used fluorescence activated cell sorting, GE profiling, and statistical modeling to generate a B-cell associated gene signature (BAGS) classifier for pre-BI, pre-BII, immature, naive, memory, and plasma cells harvested from healthy bone marrow samples during cardiac surgery.
We used the Gene Expression Omnibus (GEO) to identify 8 independent CLL cohorts with available microarray data generated on compatible array platforms. Altogether, the study included 1239 patients, who were untreated at sampling for microarray analysis.
The classifier was established using regularized multinomial regression on median-centered GE profiles from the normal B cell subsets and consisted of 184 genes with 27-54 genes per subtype. Each patient was assigned to a BAGS subtype (pre-BI, pre-BII, immature, naive, memory, and plasmacell) according to the highest probability score of the classifier. The minimum probability threshold for confident classification was chosen such that 15% remained unclassified.
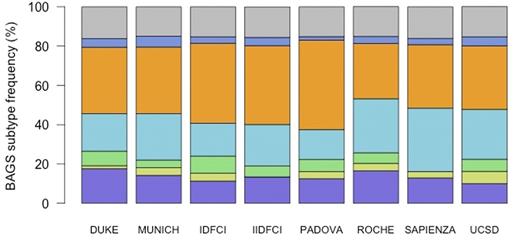
BAGS subtype frequencies in the CLL cohorts were analyzed using Fisher's exact test and revealed a distribution pattern that did not vary significantly between the 8 cohorts (P = 0.22) (Fig 1). Most samples were classified as memory (28.1-45.5%), followed by naive (15.2-32.3%), pre-BI (10.0-17.6 %), immature (0.0-8.7%), plasmacell (1.8-4.6%), and pre-BII (0.0-3.9%).
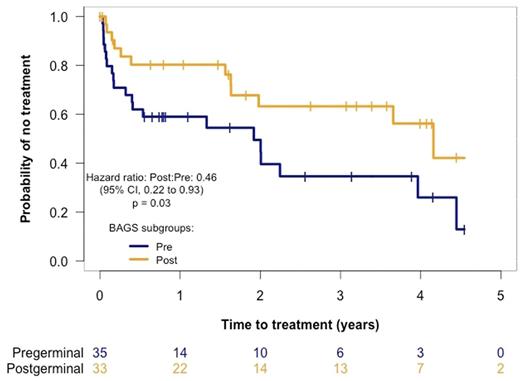
To validate the prognostic impact, our primary endpoint was time to treatment start and was evaluated using the Kaplan Meier method using available data (n = 68). We saw a significant shorter time to treatment start amongst patient with a pre-germinal center CLL subtype (pre-BI, pre-BII, immature, and naive) compared to post-germinal center subtypes (memory and plasmacell) (P = 0.03) (Fig 2).
Next, BAGS subtype was associated with the IgVH mutational status (n = 222), which showed that a larger proportion of post-germinal center subtypes had mutated IgVH genes (73.4%), whereas pre-germinal center subtypes were just as likely to have unmutated IgVH genes (48.8%) as mutated IgVH genes (52.2%).
Finally, to investigate biological differences between the BAGS subtypes, we identified differentially expressed genes (DEG) in each subtype versus the rest. Fisher's method was used to combine p-values for statistically significant DEGs in a metaanalyis including all cohorts. Of known therapeutic CLL targets, CD20 and CD52 were both significantly upregulated in the memory BAGS subtype compared to the rest.
Using a phenotypic cell of origin approach, our findings show that CLL samples display end-stage phenotypes resembling the whole spectrum of normal B-cells and indicate transcriptional plasticity as a trait that may contribute to the heterogeneity observed in CLL.
El-Galaly:Roche: Consultancy, Other: travel funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal