Abstract
Introduction: Alterations of TP53 (cytogenetic 17p13.1 deletions and molecular TP53 mutations) were reported to be frequent in pts with myelodysplastic syndromes (MDS) and complex abnormalities (≥3 clonal cytogenetic aberrations, CA) that represent around 15% of all MDS cases. It was suggested that pts with MDS and complex abnormalities may be further prognostically subdivided by the molecular TP53 mutation status (Bejar et al, ASH, 2014, abstract #532). In this study we investigated the frequency of different types of TP53 alterations, their cytogenetic profile and their clinical impact in the adverse cytogenetic MDS subgroup of complex abnormalities. We performed comprehensive cytogenetic and molecular genetic analysis focusing as well on the extent of cytogenetic instability.
Methods: We included 105 pts (57 m/48 f; median 71 yrs, range, 47-95 yrs) with MDS (n=86) and sAML after MDS (n=19) with complex abnormalities in our study. A total of 56/89 (62.9%) pts had received azacitidine. Survival was censored at allogeneic stem cell transplantation (26/83; 31.3%). Pts were characterized by chromosome banding analysis, interphasefluorescence in situ hybridization (FISH) with a panel including a 17p13/TP53-covering probe, multicolor FISH (mFISH), Sanger sequencing of TP53 and SNP-array analysis (SNP-A). The extent of genetic imbalances was objectified by counting the number of CA, the number of cytogenetic fusions as shown by mFISH and the size of total genomic aberrations (TGA) measured by SNP-A in megabases (Mb).
Results: A molecular TP53 mutation was found in 46/105 (43.8%) pts; a cytogenetic TP53 deletion in 38/105 (36.2%) pts. TP53 was not affected by a molecular mutation or a cytogenetic deletion in 44/105 (42.2%) pts, 23/105 (21.9%) pts were affected by combined TP53 alterations (molecular mutation and cytogenetic deletion), 23/105 (21.9%) pts by a molecular mutation only and 15/105 (14.3%) pts by a cytogenetic deletion only. The median number of CA was 6 (range, 3-41) in the entire cohort. Median overall survival for the entire cohort was 17 months.
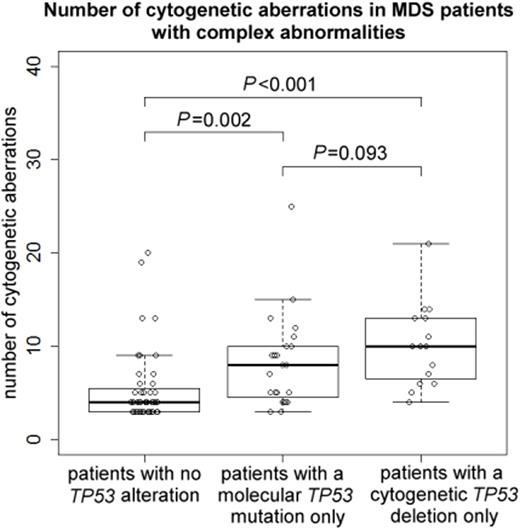
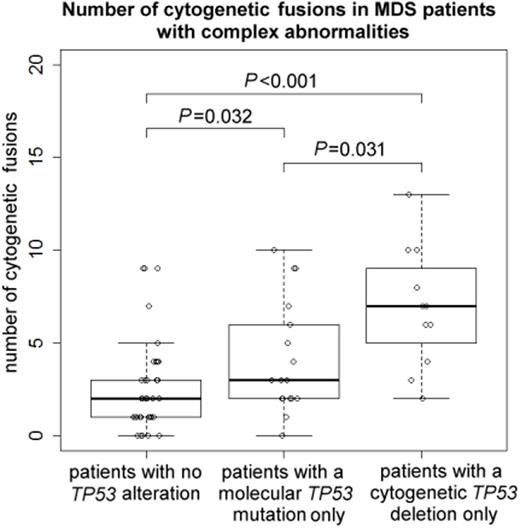
The degree of genomic imbalances was higher in pts with any TP53 alteration (molecular mutation and/or cytogenetic deletion) as compared to those without: The median number of CA was 8 (range, 3-41) vs. 4 (3-20) (P<0.001), the median number of fusions was 5 (0-13) vs. 2 (0-9) (P<0.001), and the median TGA size was 327 (97-511) vs. 105 (64-226) Mb (P<0.001). The extent of genomic imbalances was higher in pts with a TP53 deletion only compared to pts with a TP53 mutation only: The median number of CA was 10 (range, 4-21) vs. 8 (3-25) (P=0.093; n.s.); the median number of fusions was 7 (2-13) vs. 3 (0-10) (P=0.031).
By univariate analysis, presence of ≥5 CA as compared to 3-4 CA increased the hazard ratio (HR) to 3.34 (P=0.017). When we limited the analysis to the subgroup of pts without evidence of a molecular TP53 mutation, presence of a cytogenetic TP53 deletion resulted in a HR of 5.67 (P=0.029). When the analysis was restricted to pts without a TP53 deletion, presence of a molecular TP53 mutation did not significantly change the HR (1.58, P=0.448; n.s.).
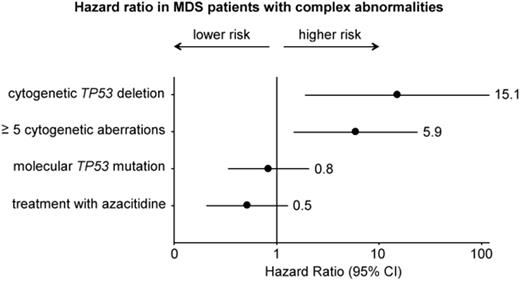
Multivariate analysis that considered molecular TP53 mutation status, cytogenetic TP53 deletion status, the number of CA and treatment with azacitidine identified presence of a cytogenetic TP53 deletion as the most significant prognostic marker for OS (HR 15.1, P=0.010). HR was increased for pts with ≥5 CA compared to pts with 3-4 CA (HR 5.9, P=0.012). The molecular TP53 mutation status showed no significant impact on HR in our cohort.
Conclusion: Presence of a cytogenetic TP53 deletion and a higher number of cytogenetic aberrations (≥5) showed a negative prognostic impact even within the unfavorable cytogenetic subgroup of MDS with complex abnormalities. In contrast, we found no strong prognostic impact for a molecular TP53 mutation in our cohort of MDS with complex abnormalities. The lower impact of TP53 mutations compared to TP53 deletions might be due to the lower degree of cytogenetic imbalances in pts with TP53 mutations only compared to pts with TP53 deletions only. However, molecular as well as cytogenetic TP53 aberrations were associated with a greater extent of chromosomal imbalances and displayed clear interdependencies. Our data suggest that TP53 alterations (molecular mutations and/or cytogenetic deletions) may not have an independent prognostic impact in the MDS subgroup with complex abnormalities.
Platzbecker:Amgen: Honoraria, Research Funding; TEVA Pharmaceutical Industries: Honoraria, Research Funding; Janssen-Cilag: Honoraria, Research Funding; Celgene Corporation: Honoraria, Research Funding; Novartis: Honoraria, Research Funding. Kroeger:Novartis: Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal