Abstract
In recent years expression levels of several genes & microRNAs (miR) were identified as strong prognostic markers, capable to refine AML risk stratification. So far technical difficulties, including the limitations of established methods for comparable, absolute quantification & the lack of defined cut points prevented translation of these findings into clinical practice. Innovative digital droplet PCR (ddPCR) is a novel technique with high sensitivity, specificity that allows absolute quantification - without the need for standard curves - & promises better inter-laboratory comparability.
In AML pts, high miR-155 expression levels associate with the presence of FLT3-ITD & independently predict inferior outcome. Here, for the first time we applied ddPCR for absolute quantification of pre-miR-155 to define an absolute cut point to discriminate between high & lowexpressers, which was then validated in a second set of AML pts.
We analyzed a homogeneous test set of 71 AML pts treated between January 2000 & June 2012 in our institution. All pts received cytarabinebased induction therapies & were consolidated with allogeneic hematopoietic stem cell transplantation (HCT) after non-myeloablativeconditioning (NMA; consisting of fludarabine[30mg / m² at days -4 to -2] & 2Gy total body irradiation [day 0]). At NMA-HCT ptswere in first (n=43; 60.6%) or second complete remission (CR; n=16; 22.5%) or CR with incomplete recovery (n=12; 16.9%). At diagnosis, cytogenetics & mutation status of the NPM1, CEBPA, IDH1, IDH2 & DNMT3A gene & presence of FLT3-ITD or FLT3-TKD mutation were assessed. The expression of the pre-miR-155 stem loop was measured using an EvaGreen-based ddPCR assay & normalized to the absolute copy numbers of ABL1.
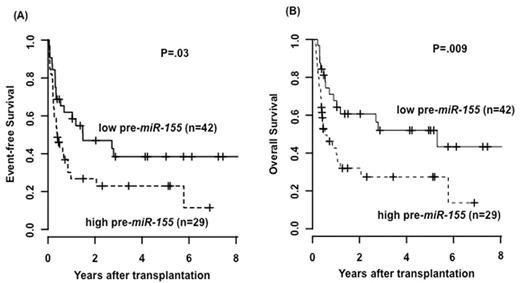
The R Package OptimalCutpointswas used to determine a cut point of 1.104 copies pre-miR-155 per 100 ABL1 copies to discriminate between high (n =29; 40.8%) & low (n =42; 59.2 %) miR-155 expressers. High miR-155 expressers, more often had a FLT3-ITD (p=.039) & less frequently a mutation in the FLT3-TKD (p=.010). No significant association was found for other clinical or biological markers at diagnosis.
In the test set, pts with more than 1.104 copies pre-miR-155 per 100 ABL1 copies at diagnosis had a significant shorter event-free survival (EFS; p=.03, figure 1A) & overall survival (OS; p=.009, figure 1B).
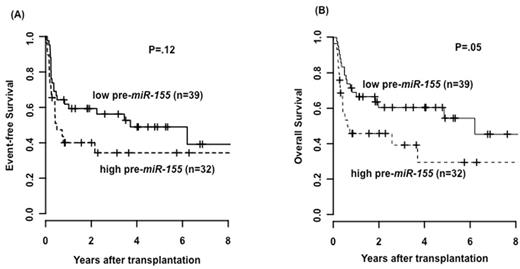
To validate these findings, we used a second set of 71 pts (median age 63.4y [range 37.1 to 74.7]) with a median follow-up of 3.7y for pts alive that all received cytarabinebased induction therapies & NMA-HCT as consolidation. The ptsin the validation set also did not differ significantly in the analyzed clinical or molecular characteristics (i.e. white blood count, hemoglobin, platelets, blasts in bone marrow or peripheral blood at diagnosis, remission status at HCT [CR1 vs. CR2 vs. CRi], ELN genetic group, mutational status of FLT3-TKD, NPM1, CEBPA, DNMT3A, IDH1 or IDH2 & presence of FLT3-ITD).
Using the determined cut point of 1.104 copies pre-miR-155 / 100 ABL1 copies in the test set, patients in the validation set were divided in 39 patients (54.9%) with a high miR-155 expression & 32 (45.1%) with a low miR-155 expression. Pts with high miR-155 expression in the validation set had shorter EFS (p=.11, figure 2A) by trend & a significant shorter OS (p=.05, figure 2B).
In conclusion, ddPCRis a novel, feasible method that allows absolute quantification of miRexpression. We defined an absolute cut point of 1.104 copies pre-miR-155 per 100 ABL1 copies for the prognosticator miR-155 in AML without the need for standard curves. Pts with pre-miR-155 expression above the cut point had a significant shorter EFS & OS. Remarkably, using a second clinically comparable set, we were able to validate our test set findings.
Future studies are planned to confirm the clinical impact of pre-miR-155 expression levels at diagnosis, as well as the identified absolute pre-miR-155 / ABL1 copy number cut point to distinguish high from low miR-155 expressers.
Test Set
Validation Test
Poenisch:Mundipharma: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal