Abstract
Background: DOCK1, also known as DOCK180, is the prototype member of the evolutionarily conserved DOCK superfamily. DOCK1 acts as a guanine nucleotide exchange factor (GEF) which regulates the activation of GTPase Rac, thereby influencing several cellular pathways including cell motility, polarity, invasion, phagocytosis and clearance of apoptotic cells. Overexpression of DOCK1 has been associated with cancer cell migration and invasion, however, its significance in acute myeloid leukemia (AML) remains unexplored.
Materials and Methods: Expression patterns of DOCK1 were extracted from 347 AML patients recruited in the National Taiwan University Hospital (NTUH) (Illumina HumanHT-12 v4 Expression BeadChip, Illumina, San Diego, CA). We employed several public datasets, including microarray data of various lineages of normal hematopoiesis and AML marrow (GSE12662, GSE24006, and GSE24759) and The Cancer Genome Atlas (TCGA) RNA sequencing data of AML. In our cohort we analyzed the impacts of DOCK1 expression on various clinical parameters and genetic abnormalities. Survival was analyzed in 227 patients who received standard chemotherapy. The global gene expression profiles associated with DOCK1 expression was analyzed by bioinformatics tools. Two independent cohorts, TCGA (N=183) and GSE12417 (N=61), were used for validating the prognostic significance of DOCK1.
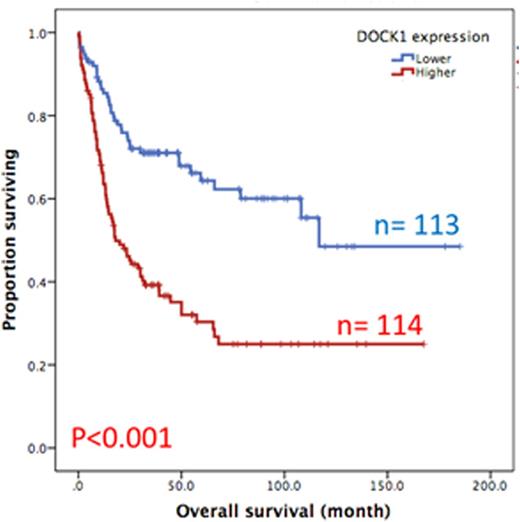
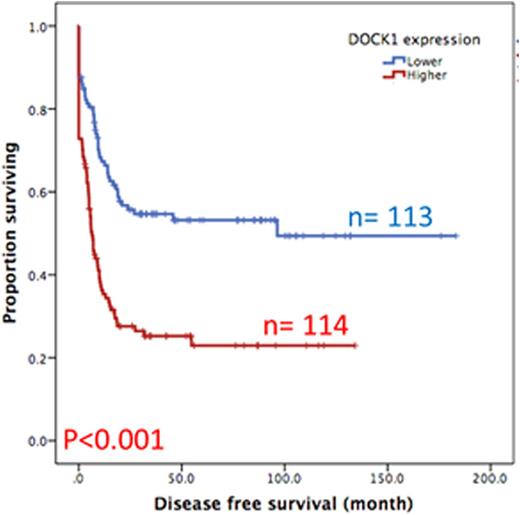
Results:DOCK1 is a moderately expressed gene in AML (RPKM, 4.2; ranking, 50.5%). Higher DOCK1 expression level was associated with higher percentages of peripheral blasts (p=0.005), but inversely correlated with favorable cytogenetics such as t(8;21) and t(15;17). Moreover, mutations in CEBPA, c-KIT and IDH2 were less often seen in the patients with higher DOCK1 expression, while FLT3-ITD, and mutations in PTPN11, MLL, NPM1, RUNX1, ASXL1 and DNMT3A happened more often in those with higher DOCK1 expression. The Gene Set Enrichment Analysis (GSEA) of published stem cell signatures suggested DOCK1 as a hematopoietic and leukemic stem cell marker (GSEA P < 0.001 and = 0.004, respectively). Furthermore, it was generally co-expressed with homeobox genes, with average correlations ranging from 0.18 to 0.33 in normal samples, and 0.10 to 0.71 in AML datasets. With a median follow up of 57 months, we observed strong correlation between higher DOCK1 expression levels and inferior overall survival (OS) (median 18.0 months vs 116.8 months, P < 0.001) and disease free survival (DFS) (median 6.4 months vs 96.4 months, P < 0.001) (Figure 1A and 1B). These survival impacts could be validated in the two independent cohorts. In addition, multivariate analysis revealed higher DOCK1 expression as a strong unfavorable prognostic factor for OS (p=0.017) independent of age, karyotype, FLT3-ITD, and mutations of PTPN11, RUNX1, MLL and TP53.
Conclusion: To our knowledge, we are the first to present the unique clinical, prognostic, and genetic impacts of DOCK1 in AML. Studies on large prospective cohorts are necessary to confirm our observation. The mechanistic functions of DOCK1 in AML await for further studies.
Overall survival of AML patients with higher expression versus lower expression of DOCK1
Overall survival of AML patients with higher expression versus lower expression of DOCK1
Disease free survival of AML patients with higher expression versus lower expression of DOCK1
Disease free survival of AML patients with higher expression versus lower expression of DOCK1
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal