Abstract
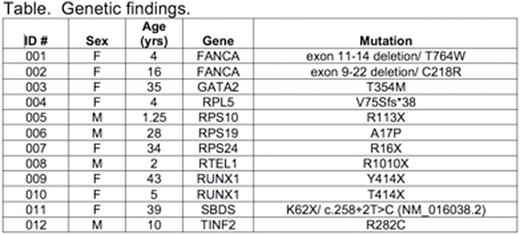
Accurate and timely diagnosis of inherited bone marrow failure (BMF) and myelodysplastic syndromes (MDS) ensures appropriate clinical management. The correct diagnosis allows appropriate monitoring for both hematopoietic (i.e. clonal evolution and progressive marrow failure) and extra-hematopoietic complications, informs the timing of hematopoietic stem cell transplant, donor selection and transplant regimen planning, and ensures appropriate genetic counseling of family members. Substantial phenotypic overlap among these disorders and the variable expressivity within syndromes complicate their diagnosis based purely on physical exam and standard laboratory testing and provide the rationale for comprehensive genetic diagnostic testing. We report here our initial one-year experience utilizing a targeted capture assay of known inherited BMF/MDS genes for clinical diagnostic purposes at the University of Washington. The assay sequences all exons and 20 base pairs of intronic sequence flanking each exon, as well as several regulatory and intronic regions of specific genes containing known pathogenic variants of 85 known inherited BMF/MDS genes (Zhang M. et al. Haematologica 2016). Between June 2015 and July 2016, 81 individual patients were referred for clinical testing (median age: 15 years-old, range: 0.6-76 years-old). For all samples evaluated, median coverage across the 383kb targeted region was 1887X. This depth of coverage enabled identification of all classes of mutations, including point mutations, small indels, copy number variants, and genomic rearrangements. Pathologic mutations in known inherited BMF/MDS genes were identified in 12 of 82 (14.6%) individuals (median age 13 years-old, range: 1.25-43 years-old) across a broad number of genes and of multiple classes including copy number variants (Table). Among the twelve patients with pathogenic mutations in inherited BMF/MDS genes, genetic testing was consistent with the prior clinical diagnoses of eight patients, including two Fanconi anemia patients subtyped as complementation group A, one of whom demonstrated reversion to wild-type resulting in mosaicism in the peripheral blood. Importantly, four patients carried no specific inherited BMF/MDS diagnosis prior to testing and were found to have pathogenic mutations in RPS10, RTEL1 and RUNX1 (ID 005, 008, 009, 010), suggesting additional diagnostic value to a multiplexed genetic approach in the clinical setting. Detailed clinical information was available for nine of the patients diagnosed with pathogenic mutations, two of whom have or will undergo a sibling or haploidentical hematopoietic stem cell transplantation (009 and 012, respectively) and thus genetic testing informed donor selection. To improve diagnostic accuracy, we are now updating the capture design to include newly discovered inherited BMF/MDS genes and intronic regions to optimize copy number variant detection. We are additionally pursuing CLIA-certified RNA analyses to characterize whether several variants bioinformatically predicted to affect splicing are functionally deleterious. Next-generation sequencing for mutations involved in hereditary marrow failure and MDS may also become increasingly important in the context of precision-medicine in which germline mutations are unexpectedly identified in somatic testing.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal