Abstract
Diamond Blackfan anemia (DBA) is congenital bone marrow failure syndrome characterized by red cell aplasia. It was the first disease linked to ribosomal dysfunction with 70% of patients having haploinsufficiency of a ribosomal protein (RP) gene. More recently, several patients with DBA have been shown to have mutations in GATA-1. The 5q- syndrome, a subtype of myelodysplastic syndrome (MDS), is also characterized by a severe anemia that is caused by heterozygous loss of RPS14 on chromosome 5q. The reciprocal relationship between these two diseases, which are now collectively known as the ribosomopathies, has spurred interest in how we may be able to better understand the pathophysiology of these disorders to broaden therapeutic options, and to improve diagnosis.
Erythrocyte adenosine deaminase (eADA) levels have been used for the last three decades in the diagnosis of DBA based on the finding in 1983 that eADA enzyme levels are significantly elevated in patients with DBA. It is now known that more than 75% of DBA patients have elevated eADA levels. The goal of the present study is to measure the levels of eADA in patients with the 5q- syndrome and determine whether they are elevated implying that eADA may be specific to ribosomal haploinsufficiency in general or normal suggesting that elevated eADA may be specific to DBA.
Under a Stanford Univeristy approved IRB protocol, we measured eADA levels in patients with the 5q- syndrome who were undergoing routine blood draws for clinical purposes. The patients with Diamond Blackfan anemia are either followed at our center in Stanford University or were previously referred to us for eADA testing with accompanying clinical information. Analysis was performed using SAS 9.3 (SAS Institute, Carey, NC). Continuous variables were presented as means and standard deviations (SD) and analyzed using 2-tailed T-Test.
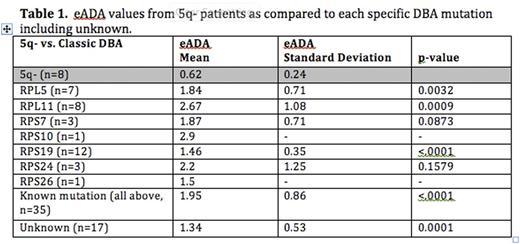
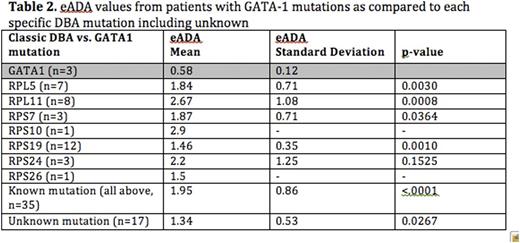
Complete results by mutation type are summarized in Tables 1 and 2.
In a disorder with the marked genotypic and phenotypic variability such as Diamond Blackfan anemia (DBA), it is critical to have sensitive tests to aid with the diagnosis. Erythrocyte adenosine deaminase (eADA) levels have long been used as supportive diagnostic criteria. The test has been especially useful in distinguishing DBA from other causes of anemia and other inherited bone marrow failure syndromes. However, the reason for the elevation of eADA in DBA remains unknown.
Our goal was to study eADA levels in the 5q- syndrome where there is an acquired ribosomal haploinsufficiency (from loss of RPS14 on chromosome 5q), which is in contrast to the congenital haploinsufficiency seen in DBA. Interestingly, we found that the eADA levels were normal in the 5q- syndrome suggesting that eADA is specific for DBA. We then studied eADA levels in patients with DBA that is not the results of a ribosomal protein (RP) mutation but instead due to mutations in GATA-1. Once again, we found that eADA levels were normal in this cohort suggesting that elevated eADA is specific feature of DBA due to RP mutations. It is interesting to note that patients with RPS19 mutations, who tend to have a milder clinical phenotype, and patients without an identified mutation had lower (but still elevated) levels of eADA when compared to other RP mutations, in particular RPL11. A larger number of patient samples in future studies would improve the statistical power to compare eADA levels between individual RP mutations.
This is the first report to examine the value of eADA in non-classical DBA and in another disorder of ribosomal haploinsufficiency, namely del(5q) MDS. In summary, we found that an elevated eADA strongly suggests the diagnosis of classic DBA although a normal eADA does not exclude the diagnosis, particularly in the setting of GATA-1 mutations. There is no utility for using eADA in the diagnosis of the 5q- syndrome despite the connection of the disease with impaired ribosome function. The reason for elevated eADA in classic DBA remains to be defined.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal