Key Points
Bacterial sepsis from contaminated platelet transfusions continues to occur despite recent interventions; additional measures are needed.
STR to platelet transfusion is frequently not recognized or reported; use of recent AABB criteria showed highest diagnostic sensitivity.
Abstract
Septic transfusion reactions (STRs) resulting from transfusion of bacterially contaminated platelets are a major hazard of platelet transfusion despite recent interventions. Active and passive surveillance for bacterially contaminated platelets was performed over 7 years (2007-2013) by culture of platelet aliquots at time of transfusion and review of reported transfusion reactions. All platelet units had been cultured 24 hours after collection and released as negative. Five sets of STR criteria were evaluated, including recent AABB criteria; sensitivity and specificity of these criteria, as well as detection by active and passive surveillance, were determined. Twenty of 51 440 platelet units transfused (0.004%; 389 per million) were bacterially contaminated by active surveillance and resulted in 5 STRs occurring 9 to 24 hours posttransfusion; none of these STRs had been reported by passive surveillance. STR occurred only in neutropenic patients transfused with high bacterial loads. A total of 284 transfusion reactions (0.55%) were reported by passive surveillance. None of these patients had received contaminated platelets. However, 6 to 93 (2.1%-32.7%) of these 284 reactions met 1 or more STR criteria, and sensitivity of STR criteria varied from 5.1% to 45.5%. These results document the continued occurrence of bacterial contamination of platelets resulting in STR in neutropenic patients, failure of passive surveillance to detect STR, and lack of specificity of STR criteria. These findings highlight the limitations of reported national STR data based on passive surveillance and the need to implement further measures to address this problem such as secondary testing or use of pathogen reduction technologies.
Introduction
Platelet transfusions are important in the prevention or treatment of bleeding in patients with thrombocytopenia or impaired platelet function.1 Approximately 2.2 million platelet transfusions were administered in the United States in 2011, with 2 million in the form of apheresis units and 200 000 as pooled whole-blood–derived units.2 Bacterial contamination of platelets is currently the leading infectious risk of platelet transfusion therapy despite recent interventions to reduce this risk.3 Over the 5-year period from 2009 to 2013, 13 fatalities associated with bacterial contamination of platelet products were recorded by the US Food and Drug Administration (FDA), a rate of 2.6 per year or ∼1.3 per million platelet transfusions.4 Regrettably, this situation is underappreciated or underrecognized by health care providers caring for patients receiving platelets, as well as by regulatory agencies, which have not yet mandated available additional measures to address this problem.5,6
Further confounding our understanding of this problem is the recognition of septic transfusion reactions (STRs) resulting from transfusion of bacterially contaminated platelets.7 Criteria for the diagnosis of STRs caused by bacterial contamination of platelets have been established to trigger recognition, management, and further investigation.8 These criteria, however, broadly overlap with the diagnostic criteria for other noninfectious transfusion reactions such as febrile-like/febrile nonhemolytic transfusion reactions (FNHTRs) and hypotensive transfusion reactions. Moreover, it has been estimated that reported STR rates have been underestimated by as much as 10-fold due to limitations of passive reporting mechanisms.9
In this study, we retrospectively evaluated the records of patients who received bacterially contaminated platelet transfusions at our institution over 7 years, as well as the records of patients with reported transfusion reactions. We analyzed the associations between bacterial species, bacterial load, and transfusion reactions, as well as the sensitivity and specificity of various STR criteria in identifying patients with true transfusion-transmitted bacterial contamination of platelets.
Patients and methods
Patients
The study was conducted at University Hospitals Case Medical Center (UHCMC), a 947-bed tertiary care academic medical center affiliated with Case Western Reserve University. Patients receiving prepooled, whole-blood–derived, random-donor platelet units (usually in pools of 5 units), and single-donor apheresis platelet units from January 1, 2007 through December 31, 2013 comprised the study population. At our institution, 75% to 80% of platelets are transfused in inpatients and 20% to 25% in outpatients. H.H., W.X., C.E.G., and R.W.M. extracted and analyzed the data from clinical and other records, and all authors had access to primary clinical data.
Platelet units
Leukoreduced apheresis and prepooled whole-blood–derived platelet units were obtained primarily from local blood suppliers, the American Red Cross (ARC) of Northeast Ohio (Cleveland, OH), and LifeShare Community Blood Services (Elyria, OH). Both suppliers diverted the first 5 to 10 mL of each collection, cultured all platelet units 24 hours after collection, and released units if culture was negative 24 hours later. Culture was performed by inoculation of 8 to 10 mL of platelets into BacT/ALERT BPA culture bottles (ARC) or 3 to 4 mL into eBDS culture pouches (LifeShare).
Surveillance for bacterial contamination of platelet units and STRs
Active and passive surveillance were performed on all platelet products issued and patients receiving platelet products during the study period.7 Active surveillance was performed by collecting 1- to 2-mL aliquots of apheresis or pooled platelet units at the time of issue for culture (see “Microbiology methods” below). Patients were evaluated for evidence of transfusion reactions as soon as positive culture results were obtained. Passive surveillance in our institution depends on reporting of transfusion reactions by clinicians in outpatient and inpatient settings to the transfusion medicine service; no institutional transfusion safety officer has been appointed. Detailed instructions (kept updated with the most recent FDA hemovigilance guidelines, providing guidance on diagnosis, timing of reporting, laboratory workup, sample collection, and treatment according to the type of transfusion reaction encountered) have been developed and are available on the hospital’s intranet service. Additionally, these instructions include educational materials and forms that are required to accompany all samples sent to the blood bank for testing. Further assistance is provided by the transfusion medicine service once notification has occurred. All reactions are then investigated by the transfusion medicine service, which follows up the patient’s clinical status to determine whether resolution of the reaction occurred following treatment and analyzes applicable laboratory results. Transfusion reaction investigations are not considered complete until a transfusion medicine physician writes a comprehensive note verifying that the investigation has been completed and determines that all appropriate testing has been performed. Investigation of transfusion reactions to platelets includes patient evaluation and Gram stain and culture of the remains of implicated platelet units. Patients’ transfusion reactions, documented by active or passive surveillance, were classified by severity as previously described7 and correlated with platelet and posttransfusion blood cultures and other investigations.
Microbiology methods
For active surveillance, bacterial culture was performed by plating 0.1-mL aliquots of platelet samples obtained at time of issue onto blood agar plates, which were incubated for up to 48 hours in 5% CO2 at 35°C; isolates recovered were identified, and antimicrobial susceptibility testing performed according to standard procedures, and preserved at −70°C.10 Culture was repeated on platelet specimens with initial positive culture results from samples retained at 4°C to determine whether results were true or false positives. True positives were defined as isolation of the same bacterial species from the repeat culture. Quantitative cultures to determine bacterial load were also performed on initial positive cultures by plating 0.1 mL of serial 10-fold dilutions of platelet specimens (retained at 4°C to prevent further bacterial replication) onto blood agar plates, which were incubated aerobically for up to 48 hours in 5% CO2 at 35°C. Culture results were recorded as colony-forming units (cfu) per mL of platelet product. Bacterial loads were interpreted as high if ≥105 cfu/mL and low if <105 cfu/mL, as previously described.7
For passive surveillance, Gram stain and plate culture of the remains of implicated platelet units were performed. The culture method was the same as used for active surveillance with the addition of a second blood agar plate, which was incubated anaerobically for up to 48 hours at 35°C.
Blood cultures on patients were performed according to standard procedures using SA aerobic and SN anaerobic blood culture bottles (BacT/ALERT; Biomerieux).
STR criteria
The records of patients who received bacterially contaminated platelets (ie, detected by active surveillance) as well as those with reported transfusion reactions after platelet transfusion (ie, detected by passive surveillance) were evaluated and reactions classified according to reaction type as previously described.7,11 Reactions were also classified using 5 sets of STR criteria, including those from the Centers for Disease Control and Prevention (CDC) Biovigilance Guidelines12 and recommendations from Sanders et al/St. Jude Children’s Research Hospital (SJCRH),13 ARC,8,14 Blood Safety Surveillance and Health Care Acquired Infections Division, Public Health Agency of Canada (PHAC),15 and AABB5 (Table 1). These criteria are based on the clinical features of transfusion reactions, including symptoms, observation period for onset of symptoms after platelet transfusion, and changes in vital signs. The length of the observation period for onset of symptoms after transfusion in these criteria varies from 4 to 24 hours. Patient records were also reviewed for evidence of delayed sequela.16
STR definitions and criteria used to analyze study findings
| STR criterion . | STR definition (observation period) . | ||||
|---|---|---|---|---|---|
| CDC11 (4 h) . | SJCRH4 (24 h) . | ARC8,12 (4 h) . | PHAC13 (6 h) . | AABB6 (24 h) . | |
| A | Fever ≥39°C | Fever ≥40°C | Fever ≥39°C | Fever ≥39°C with a rise of ≥1°C | Fever ≥38°C with a rise of ≥1°C PLUS any of: rigors, dyspnea, nausea/vomiting, tachycardia, hypotension, shock |
| B | A rise of ≥2°C PLUS any of: rigors, dyspnea, nausea/vomiting, tachycardia, hypotension, shock | Cardiovascular collapse | A rise of ≥2°C | Fever ≥38°C with a rise of ≥1°C PLUS any of: rigors, dyspnea, nausea/vomiting, tachycardia, hypotension, shock | Any sepsis like syncope/hypotension |
| C | Any of: rigors, tachycardia, hypotension, shock | Any other suspicious clinical changes | |||
| STR criterion . | STR definition (observation period) . | ||||
|---|---|---|---|---|---|
| CDC11 (4 h) . | SJCRH4 (24 h) . | ARC8,12 (4 h) . | PHAC13 (6 h) . | AABB6 (24 h) . | |
| A | Fever ≥39°C | Fever ≥40°C | Fever ≥39°C | Fever ≥39°C with a rise of ≥1°C | Fever ≥38°C with a rise of ≥1°C PLUS any of: rigors, dyspnea, nausea/vomiting, tachycardia, hypotension, shock |
| B | A rise of ≥2°C PLUS any of: rigors, dyspnea, nausea/vomiting, tachycardia, hypotension, shock | Cardiovascular collapse | A rise of ≥2°C | Fever ≥38°C with a rise of ≥1°C PLUS any of: rigors, dyspnea, nausea/vomiting, tachycardia, hypotension, shock | Any sepsis like syncope/hypotension |
| C | Any of: rigors, tachycardia, hypotension, shock | Any other suspicious clinical changes | |||
Observation period refers to the time period following transfusion during which changes in signs and symptoms should be regarded as associated with the transfusion. Patients meeting any criterion (A, B, or C) during the observation period are deemed to have STR.
Data sources
After approval from the UHCMC Institutional Review Board, data were extracted from available sources, including surveillance records, clinical charts, blood bank investigation logs, transfusion reaction reports, and laboratory records.
Statistical analysis
All statistics were performed using Prism 6 (GraphPad Software Inc). Results are presented as mean ± standard deviation. The intergroup data comparisons were performed using the Student t test or χ2 test. All reported P values are 2-sided with a type I error rate of 5% and a P < .05 set for significance.
Results
A total of 51 440 leukoreduced platelet products were transfused during the 7-year study period, of which 38 692 (75.2%) were apheresis units and 12 748 (24.8%) were pooled units. A total of 284 transfusion reactions were reported by passive surveillance during the observation period. The rate of transfusion reactions was not significantly different between inpatients and outpatients.
Bacterial culture results of platelet units
Twenty of the 51 440 platelet products (389 per million) transfused during the study period were detected as bacterially contaminated by active surveillance. The incidence of bacterial contamination was 4 of 12 748 (314 per million) in prepooled products and 16 of 38 692 (414 per million) in apheresis units (χ2 test, P = .62). Contaminants included 19 gram-positive species, predominantly coagulase-negative staphylococci (n = 13) and streptococci (n = 5), and only 1 gram-negative species, Acinetobacter baumannii (Table 2). Two pairs of isolates, Staphylococcus epidermidis (cases 56 and 57) and Streptococcus gallolyticus (Streptococcus bovis) (cases 64 and 65), were from split apheresis units from the same collections. Bacterial loads ranged from 4 × 102 cfu/mL to 6 × 107 cfu/mL, with 11 units having loads of ≥105 cfu/mL. Demographic information on the 20 patients who received bacterially contaminated platelet products is shown in supplemental Table 1 (see supplemental Data, available on the Blood Web site).
Bacterial culture results of bacterially contaminated platelets administered to patients, with patient peripheral blood leukocyte count, transfusion reactions, and posttransfusion blood culture results
| Case no. . | Platelet type . | Organism . | WBC, 103/μL . | Load, cfu/mL . | Transfusion reaction . | Posttransfusion blood culture . |
|---|---|---|---|---|---|---|
| 55 | Prepooled | S epidermidis | 9.4 | 2.0 × 104 | No | ND |
| 56 | Apheresis | S epidermidis | 0.7 | 6.0 × 106 | No | Negative |
| 57 | Apheresis | S epidermidis | 27.7 | 3.0 × 106 | No | Negative |
| 58 | Apheresis | Staphylococcus warneri | 0.3 | 2.2 × 102 | No | Negative |
| 59 | Apheresis | S epidermidis | 16.9 | 1.5 × 107 | No | ND |
| 60 | Apheresis | A baumannii | 2.1 | 4.0 × 102 | No | Negative |
| 61 | Apheresis | S epidermidis | 4.1 | 1.0 × 106 | No | Negative |
| 62 | Apheresis | Streptococcus sanguinis | 0.1 | 6.0 × 107 | No | Negative |
| 63 | Prepooled | S aureus | 1.4 | 1.4 × 105 | Yes | Positive* |
| 64 | Apheresis | S gallolyticus | 3 | 1.0 × 102 | No | Negative |
| 65 | Apheresis | S gallolyticus | 6.8 | 1.0 × 102 | No | Negative |
| 66 | Apheresis | S epidermidis | 11 | 8.5 × 103 | No | Negative |
| 67 | Prepooled | S epidermidis | 10.8 | 1.0 × 105 | No | Negative |
| 68 | Apheresis | S warneri | 0.5 | 5.0 × 106 | Yes | Positive* |
| 70 | Apheresis | S oralis | 0.7 | 7.0 × 106 | Yes | Positive* |
| 71 | Apheresis | S epidermidis | 7.4 | 4.0 × 102 | No | Negative |
| 72 | Apheresis | S epidermidis | 2.6 | 5.0 × 106 | Yes | Negative |
| 74 | Prepooled | S epidermidis | 3.9 | 5.0 × 104 | No | Negative |
| 75 | Apheresis | S epidermidis | 10.8 | 4.0 × 102 | No | ND |
| 76 | Apheresis | S oralis | 1.1 | 1.1 × 106 | Yes | Positive* |
| Case no. . | Platelet type . | Organism . | WBC, 103/μL . | Load, cfu/mL . | Transfusion reaction . | Posttransfusion blood culture . |
|---|---|---|---|---|---|---|
| 55 | Prepooled | S epidermidis | 9.4 | 2.0 × 104 | No | ND |
| 56 | Apheresis | S epidermidis | 0.7 | 6.0 × 106 | No | Negative |
| 57 | Apheresis | S epidermidis | 27.7 | 3.0 × 106 | No | Negative |
| 58 | Apheresis | Staphylococcus warneri | 0.3 | 2.2 × 102 | No | Negative |
| 59 | Apheresis | S epidermidis | 16.9 | 1.5 × 107 | No | ND |
| 60 | Apheresis | A baumannii | 2.1 | 4.0 × 102 | No | Negative |
| 61 | Apheresis | S epidermidis | 4.1 | 1.0 × 106 | No | Negative |
| 62 | Apheresis | Streptococcus sanguinis | 0.1 | 6.0 × 107 | No | Negative |
| 63 | Prepooled | S aureus | 1.4 | 1.4 × 105 | Yes | Positive* |
| 64 | Apheresis | S gallolyticus | 3 | 1.0 × 102 | No | Negative |
| 65 | Apheresis | S gallolyticus | 6.8 | 1.0 × 102 | No | Negative |
| 66 | Apheresis | S epidermidis | 11 | 8.5 × 103 | No | Negative |
| 67 | Prepooled | S epidermidis | 10.8 | 1.0 × 105 | No | Negative |
| 68 | Apheresis | S warneri | 0.5 | 5.0 × 106 | Yes | Positive* |
| 70 | Apheresis | S oralis | 0.7 | 7.0 × 106 | Yes | Positive* |
| 71 | Apheresis | S epidermidis | 7.4 | 4.0 × 102 | No | Negative |
| 72 | Apheresis | S epidermidis | 2.6 | 5.0 × 106 | Yes | Negative |
| 74 | Prepooled | S epidermidis | 3.9 | 5.0 × 104 | No | Negative |
| 75 | Apheresis | S epidermidis | 10.8 | 4.0 × 102 | No | ND |
| 76 | Apheresis | S oralis | 1.1 | 1.1 × 106 | Yes | Positive* |
ND, not done.
Same organism as in platelet product.
No additional contaminated units were detected among the 284 units recultured following reported transfusion reactions (passive surveillance), which included febrile-like/FNHTR, allergic, and other transfusion reactions.
STR in patients receiving bacterially contaminated platelet units
Ten patients who received contaminated platelets had hematologic malignancies: 2 had other malignancies and 8 had nonneoplastic thrombocytopenia (supplemental Table 1). Four were undergoing hematopoietic stem cell transplants. None of the patients were receiving antimicrobial therapy on day of transfusion. Five of the 20 patients who received contaminated platelets developed signs and symptoms that should have been reported to the transfusion service as transfusion reactions according to transfusion policies (Table 3). However, none of these reactions had been reported to the transfusion service and were only revealed during investigation of positive platelet cultures. Four of these patients received their transfusions in outpatient clinics and the fifth was an inpatient at the time of transfusion. These 5 patients had no symptoms or signs of sepsis prior to transfusion. All 5 patients had delayed-onset reactions, occurring 9 to 24 hours posttransfusion. Transfusion reactions were recognized and patients treated appropriately in 3 of these cases (all outpatients). The reaction in another outpatient was only recognized when the positive culture was reported and the patient was called in for treatment. The remaining reaction occurred in an inpatient, where the patient presented in cardiac arrest. Bacterial species involved were Staphylococcus aureus (n = 1), coagulase-negative staphylococci (n = 2), and Streptococcus oralis (n = 2). Posttransfusion blood culture yielded the same bacterial species as detected in the product in 4 of the 5 patients. Of the 15 patients who had no reaction, 13 had blood cultures performed within 48 hours of receipt of contaminated platelets which were negative; the remaining 2 patients did not have posttransfusion blood cultures performed within this time period.
Clinical presentations of patients with confirmed STR
| Case no. . | Age, y/sex . | Diagnosis . | In/out patient . | Onset, h . | Presentation . | Severity . |
|---|---|---|---|---|---|---|
| 63 | 56/M | MM PBSCT | Out | 24 | Hypotension syncope | Moderate |
| 68 | 62/M | AML BMT | In | 12 | Cardiac arrest multiorgan failure | Fatal |
| 70 | 78/M | NHL | Out | 9 | Fever (39.5°C), rigors | Life-threatening |
| 72* | 22/F | AA | Out | 18 | Hypotension | Moderate |
| 76 | 7/M | ALL | Out | 16 | Fever (39.5°C), rigors | Severe |
| Case no. . | Age, y/sex . | Diagnosis . | In/out patient . | Onset, h . | Presentation . | Severity . |
|---|---|---|---|---|---|---|
| 63 | 56/M | MM PBSCT | Out | 24 | Hypotension syncope | Moderate |
| 68 | 62/M | AML BMT | In | 12 | Cardiac arrest multiorgan failure | Fatal |
| 70 | 78/M | NHL | Out | 9 | Fever (39.5°C), rigors | Life-threatening |
| 72* | 22/F | AA | Out | 18 | Hypotension | Moderate |
| 76 | 7/M | ALL | Out | 16 | Fever (39.5°C), rigors | Severe |
AA, aplastic anemia; ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; BMT, bone marrow transplant; MM, multiple myeloma; NHL, non-Hodgkin lymphoma; PBSCT, peripheral blood stem cell transplant.
Posttransfusion blood culture was negative.
The clinical presentations of the 5 patients with STR included fever/rigors (n = 2), hypotension (n = 1), hypotension and fever (n = 1), and cardiac arrest (n = 1) (Table 3). These reactions were not initially identified as STR, but were recorded as FNHTR or hypotensive transfusion reactions in patients’ medical records. After chart review, communication with clinical teams, and correlation with culture results, these 5 reactions were clinically deemed to be STR: 1 fatal, 1 life-threatening, 1 severe, and 2 moderate. In the fatal case, the patient developed cardiac arrest 12 hours posttransfusion, which progressed to multiorgan failure, and the patient died 10 days later. No other adverse events or delayed sequela to receipt of contaminated platelets such as central venous line infections with the same bacterial species present in contaminated units were detected on review of long-term patient records.
Bacterial load and leukocyte count at time of transfusion in patients transfused with bacterially contaminated platelets
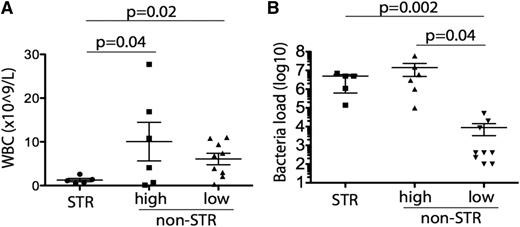
Bacterial loads were high (≥105 cfu/mL) in 11 of the contaminated units transfused and low (<105 cfu/mL) in the other 9 transfused units (Table 2). Of the 11 patients transfused with high bacterial load products, 5 developed STR. In contrast, none of the 9 patients receiving low bacterial loads developed STR (Figure 1A; supplemental Table 2). The median bacterial load was 2 log10 higher in products transfused to patients who developed STR than that in products not resulting in transfusion reactions (5 × 106 vs 2 × 104 cfu/mL, P = .005) (Figure 1B). Leukocyte counts at time of transfusion were eightfold lower in the 5 patients who had STR than in the 6 who did not but were transfused with high-load contaminated products (1.26 ± 0.37 × 109/L vs 10.1 ± 4.4 × 109/L, P = .04 by t test) (Figure 1A).
Relationships between WBC and STR and bacterial load and STR. Correlation of WBC count (A) and bacterial load (B) in patients who received bacterially contaminated platelets with presence or absence of STR following transfusion. Bacterial load is shown in cfu/mL. Of the 11 patients who received platelet units with high bacterial loads, 5 developed STRs whereas 6 did not; none of the 9 patients who received platelet units with low bacterial loads developed STRs. STRs were significantly associated with low WBC count (A) and high bacterial loads (B).
Relationships between WBC and STR and bacterial load and STR. Correlation of WBC count (A) and bacterial load (B) in patients who received bacterially contaminated platelets with presence or absence of STR following transfusion. Bacterial load is shown in cfu/mL. Of the 11 patients who received platelet units with high bacterial loads, 5 developed STRs whereas 6 did not; none of the 9 patients who received platelet units with low bacterial loads developed STRs. STRs were significantly associated with low WBC count (A) and high bacterial loads (B).
Sensitivity and specificity of active vs passive surveillance in detecting any transfusion reaction
As noted under “STR in patients receiving bacterially contaminated platelet units,” no reactions were reported from the 20 patients who received contaminated platelets, although reactions had occurred and had been documented in patients’ medical records in 5 cases, whereas 284 reactions were reported on patients who received sterile platelets. Demographic information on the 284 patients who did not receive bacterially contaminated platelet products was similar to that of patients who received contaminated platelet products and is shown in supplemental Table 3. Although the specificity of passive surveillance to detect transfusion of bacterially contaminated platelets was high (51 156 of 51 420, 99.5%), sensitivity was zero (0 of 20) (supplemental Table 4). In contrast, sensitivity of active surveillance was 25% (5 of 20), whereas specificity was the same as that for passive surveillance.
Sensitivity and specificity of STR criteria
The sensitivity and efficiency of the diagnostic criteria currently available for detecting STR was determined for the 5 patients who developed reactions after receiving bacterially contaminated platelets. All 5 patients met AABB criteria, whereas only 3 met SJCRH criteria and 2 PHAC criteria; none met CDC or ARC criteria (Table 4). The single most important reason for patients not meeting STR criteria was the short observation time of 4 to 6 hours used by CDC, ARC, and PHAC (Table 1). For SJCRH criteria, the requirement for fever of ≥40°C resulted in 2 patients not meeting criteria. The AABB criteria therefore showed the highest sensitivity (100%) whereas the sensitivities of the CDC, ARC, SJCRH, and PHAC criteria varied from 0% to 60%.
Number of patients with clinically confirmed STR meeting various STR criteria
| Total TR (n = 5) . | CDC . | SJCRH . | ARC . | PHAC . | AABB . |
|---|---|---|---|---|---|
| Cases meeting STR criteria | 0 | 3 | 0 | 2 | 5 |
| Reasons for not meeting various STR criteria | |||||
| Long latency (>6 h) | 5 | 0 | 5 | 3 | 0 |
| Temperature did not reach ≥40°C | 0 | 2 | 0 | 0 | 0 |
| Total TR (n = 5) . | CDC . | SJCRH . | ARC . | PHAC . | AABB . |
|---|---|---|---|---|---|
| Cases meeting STR criteria | 0 | 3 | 0 | 2 | 5 |
| Reasons for not meeting various STR criteria | |||||
| Long latency (>6 h) | 5 | 0 | 5 | 3 | 0 |
| Temperature did not reach ≥40°C | 0 | 2 | 0 | 0 | 0 |
To examine the specificity of these STR criteria, we analyzed reported transfusion reactions in patients receiving culture-negative platelet products (n = 51 420). A total of 284 reactions were reported (incidence: 0.55%; apheresis 199 of 38 675 [0.51%] vs prepooled 85 of 12 744 [0.67%]), with similar rates of reaction types to apheresis and prepooled units. The demographic information for these patients was also similar to that of patients who received contaminated platelets (supplemental Table 3). Of these reactions, 127 (44.7%) were categorized as FNHTR/febrile-like transfusion reactions (TR), 111 (39.1%) as allergic TR, and 46 (16.2%) as other TR (Table 5). Overall, 36 of these 284 reactions met AABB STR criteria, 52 met PHAC criteria, and 93 met CDC and ARC criteria; only 6 met SJCRH criteria (Table 6). Of 127 patients with FNHTR/febrile-like transfusion reactions, 30 met AABB STR criteria, 48 PHAC criteria, and 81 CDC and ARC criteria; none met SJCRH criteria. Of 46 other transfusion reactions, 6 met AABB STR criteria, 4 PHAC criteria, 12 CDC and ARC criteria, and 6 SJCRH criteria. None of 111 allergic TR met any of the STR criteria.
Details of 284 transfusion reactions in patients receiving 51 420 culture-negative platelet units from 2007 to 2013
| Reaction type . | No. . | Reaction rate, % . |
|---|---|---|
| Febrile/FNHTR | 127 | 0.29 |
| Allergic transfusion reaction | 111 | 0.26 |
| Other | 46 | 0.11 |
| Total | 284 | 0.66 |
| Reaction type . | No. . | Reaction rate, % . |
|---|---|---|
| Febrile/FNHTR | 127 | 0.29 |
| Allergic transfusion reaction | 111 | 0.26 |
| Other | 46 | 0.11 |
| Total | 284 | 0.66 |
Numbers of patients with transfusion reactions meeting various STR criteria who received culture-negative platelets (n = 51 420) according to reaction type
| Reaction type . | Cases meeting STR criteria . | ||||
|---|---|---|---|---|---|
| CDC11 . | SJCRH4 . | ARC8,12 . | PHAC13 . | AABB6 . | |
| Allergic TR (n = 111) | 0 | 0 | 0 | 0 | 0 |
| FNHTR/Febrile-like TR (n = 127) | 81 | 0 | 81 | 48 | 30 |
| Other TR (n = 46) | 12 | 6 | 12 | 4 | 6 |
| Total (n = 284) | 93 | 6 | 93 | 52 | 36 |
Sensitivity, specificity, and positive and negative predictive values of the 5 STR sets of criteria are shown in Table 7. Specificity and negative predictive values were high (>99%) for all STR criteria sets, but sensitivity varied from 0% to 100% and positive predictive value from 0% to 33.3%. Overall, AABB criteria provided the best predictive values. As none of the 5 STRs had been reported to the transfusion service and were only detected by active surveillance, sensitivity and positive predictive value of STRs reported by passive surveillance was 0%. Overall sensitivity of STR detection by combined active and passive surveillance varied from 5 of 98 (5.1%) to 5 of 11 (45.5%); sensitivity of AABB criteria was 5 of 41 (35.7%).
Evaluation of sensitivity, specificity, and positive and negative predictive values of 5 sets of STR criteria in identifying patients with true septic reactions
| . | Results of analysis using specified STR criteria (% values with 95% CI in parentheses) . | ||||
|---|---|---|---|---|---|
| CDC . | SJCRH . | ARC . | PHAC . | AABB . | |
| Sensitivity (n = 5) | 0 (0-52.2) | 60 (14.7-94.7) | 0 (0-52.2) | 40 (5.3-85.3) | 100 (47.8-100) |
| Specificity (excluding allergic TR) (n = 173) | 99.82 (97.78-99.85) | 99.99 (99.97-100) | 99.82 (97.78-99.85) | 99.90 (99.87-99.92) | 99.93 (99.90-99.95) |
| Positive predictive value | 0 (0-3.89) | 33.33 (7.5-70.1) | 0 (0-3.89) | 3.7 (0.45-12.75) | 12.20 (4.1-26.2) |
| Negative predictive value | 99.99 (99.98-100) | 100 (99.99-100) | 99.99 (99.98-100) | 99.99 (99.98-100) | 100 (99.99-100) |
| . | Results of analysis using specified STR criteria (% values with 95% CI in parentheses) . | ||||
|---|---|---|---|---|---|
| CDC . | SJCRH . | ARC . | PHAC . | AABB . | |
| Sensitivity (n = 5) | 0 (0-52.2) | 60 (14.7-94.7) | 0 (0-52.2) | 40 (5.3-85.3) | 100 (47.8-100) |
| Specificity (excluding allergic TR) (n = 173) | 99.82 (97.78-99.85) | 99.99 (99.97-100) | 99.82 (97.78-99.85) | 99.90 (99.87-99.92) | 99.93 (99.90-99.95) |
| Positive predictive value | 0 (0-3.89) | 33.33 (7.5-70.1) | 0 (0-3.89) | 3.7 (0.45-12.75) | 12.20 (4.1-26.2) |
| Negative predictive value | 99.99 (99.98-100) | 100 (99.99-100) | 99.99 (99.98-100) | 99.99 (99.98-100) | 100 (99.99-100) |
CI, confidence interval.
Discussion
STR is one of the most commonly recognized causes of transfusion-related fatalities following transfusion-associated acute lung injury, hemolytic TR, and transfusion-associated circulatory overload.4,5,17-20 Despite the recognition of these complications of platelet transfusion, their true incidence is unknown as a result of underdetection and underreporting, notwithstanding improved hemovigilance programs. A recent report on transfusion-associated circulatory overload showed considerable underreporting, with a 36-fold difference in detection between active and passive surveillance: 1:5997 cases reported by passive surveillance vs 1:167 by active surveillance.21 Furthermore, there is considerable variation in the reported incidence of transfusion reactions to platelets, with recent reports ranging from 0.01% to 10.0%, with considerable variation in reaction severity definitions and surveillance methods likely accounting for this wide range.22-24
Platelet components are particularly vulnerable to bacterial contamination, mainly due to their storage at room temperature, which allows growth of small inocula to very high titers and limits the shelf-life of these products.12,25 For several decades, efforts have been made at multiple levels to reduce STR and fatalities from bacterially contaminated platelet transfusions, including prevention of contamination during collection and processing,8,26 culture of platelet products 24 hours after collection,3,27-29 point-of-issue testing,27,30,31 as well as pathogen-reduction technologies.32,33 Introduction of culture of platelet products in 2004 led to a decrease in gram-negative contaminants, which had accounted for one-third of contaminants and two-thirds of fatalities.34 However, although considerable progress has been made, pathogen transmission through transfusion has continued and STRs continue to occur.6 In the United States, 13 fatalities were reported to the FDA associated with platelet transfusion-transmitted bacterial sepsis during the previous 5 fiscal years (2009-2013), with 4 associated with pooled platelets and 9 with apheresis units.4 The fatality rate associated with gram-positive sepsis in our study is similar to that reported in the literature.34
Our study highlights these issues and extends previous findings at our institution that bacterial contamination of platelets is an ongoing problem and that detection by passive surveillance continues to be poor.7,10 Specifically, none of the 5 STRs that occurred during the 7-year study period had been reported to the transfusion service, and cases were only documented as a result of our active surveillance program. This finding is similar to our experience in 1991 to 2006, where active surveillance detected 32-fold more bacterially contaminated platelet units and 10.6-fold more septic reactions than did passive surveillance.7 During this previous period, 54 bacterial contaminants included 4 gram-negative bacilli, 3 of which were associated with fatal reactions, 44 staphylococci, 4 streptococci, and 2 Bacillus cereus. This contrasts with the current study (2007-2013), in which staphylococci and streptococci accounted for 19 of 20 contaminants; the decrease in gram-negative bacilli was likely the result of introduction of early culture in 2004, with improved detection of these rapidly growing species. This decrease in virulent gram-negative contaminants is also reflected in reported national fatality data, where fatalities from gram-negative species decreased from 3.7 per year prior to introduction of early culture to 1.2 per year following use of early culture vs 2.3 and 2.0 per year, respectively, for gram-positive species.4 These findings and trends emphasize the importance of monitoring patients receiving platelet transfusions closely for at least 24 hours, investigating all reactions for STR by culture of platelet products regardless of severity of reactions, and reporting all reactions to the transfusion service. Although the yield from culture of products administered to patients who develop any reaction is low, culture or other microbiologic testing is the only practical method of differentiating STRs from other reaction types due to overlap of signs and symptoms. Use of active surveillance of patients by dedicated hemovigilance personnel has been shown to improve recognition of STRs.19 We also noted that STRs occurred with high bacterial loads only when patients were neutropenic (white blood cell [WBC] count <2 × 109/L), likely related to the relatively low virulence of the gram-positive contaminants transfused. However, we did not find any evidence of delayed sequela resulting from transfusion of contaminated platelets. In particular, no cases of central line infections caused by bacterial isolates present in contaminated platelets were found. Although central line infections did occur, causative pathogens were very different from those associated with platelet contaminants. Specifically, strains of Staphylococcus epidermidis and other coagulase-negative staphylococci associated with contaminated platelets were very susceptible to antimicrobial agents, consistent with their origin from the skin of healthy donors. In contrast, coagulase-negative staphylococci associated with central line infections were methicillin and multidrug resistant, consistent with their origin from the skin of patients exposed to antimicrobial agents and the hospital environment.
The need to take additional measures to address the problem of bacterial contamination of platelets has recently been acknowledged by the AABB and the FDA.5,6 An AABB Association Bulletin in 2012 recommended that blood establishments develop a policy or policies to further reduce the residual risk of bacterial contamination of apheresis platelets, improve the recognition of and monitoring for STRs to all platelet components, and optimize appropriate transfusion practice for all platelet components.35 The AABB subsequently issued a bulletin in 2014 emphasizing the need to recognize and provide a timely response to suspected STRs and to protect other patients from receiving contaminated co-components.5 The FDA has also recently issued a draft guidance document (Bacterial Detection Testing by Blood Collection Establishments and Transfusion Services to Enhance the Safety and Availability of Platelets for Transfusion) recommending additional testing by culture or rapid test during the storage period, or use of pathogen reduction technologies at time of production to further reduce the risk of bacterial contamination.6
As documented in this Discussion, the problems of bacterial contamination of platelets and resultant STRs are well recognized and the means to address them are available, including delayed primary testing, use of secondary testing by culture or rapid device during storage, and use of pathogen reduction systems; regulatory agencies strongly recommend use of such systems.5,6 Another issue that needs to be addressed is improving both the sensitivity and specificity of diagnosing STRs. The recent AABB criteria5 provided the best combination of sensitivity and specificity, but were compromised in our series by the fact that none of these reactions were reported to the transfusion service. Lack of reporting was, in part, associated with delayed reactions to transfusions administered in satellite outpatient clinics, with different clinical teams seeing patients in acute care settings to those administering the transfusions. Additionally, overlap in signs and symptoms of other more common TR further limits clinical diagnosis, and no specific temperature threshold or clinical feature was shown to be highly predictive of STRs.
In conclusion, our findings document the continued occurrence of bacterially contaminated platelets leading to STRs and highlight the need for implementation of additional, available measures such as secondary testing or pathogen reduction to further reduce these reactions and fatalities.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The contribution of the late Roslyn A. Yomtovian, MD, Medical Director, Transfusion Service, University Hospitals Case Medical Center and Case Western Reserve University School of Medicine, Cleveland, OH, 1988-2007, to the field of bacterial contamination of platelets is recognized with appreciation for her early recognition of this problem and her contributions to its study and prevention.
Authorship
Contribution: H.H., W.X., C.E.G., R.W.M., and M.R.J. extracted data from patient and laboratory records and analyzed results; W.X. made the figures; R.W.M. and M.R.J. designed the research; and H.H., W.X., H.M.L., C.E.G., R.W.M., and M.R.J. wrote the paper.
Conflict-of-interest disclosure: M.R.J. has received research support and/or honoraria from Verax, Pall, Gambro, Hemosystem, Immunetics, Genprime, Fenwal, and Charles River Labs, has been a consultant for BioSense Technologies and Lynntech, Inc, and is a member of the Bacterial Contamination Task Forces of the AABB and the International Society of Blood Transfusion (ISBT). The remaining authors declare no competing financial interests.
Correspondence: Michael R. Jacobs, Department of Pathology, University Hospitals Case Medical Center, 11100 Euclid Ave, Cleveland, OH 44106; e-mail: michael.jacobs@case.edu; and Robert W. Maitta, Department of Pathology, University Hospitals Case Medical Center, 11100 Euclid Ave, Cleveland, OH 44106; e-mail: robert.maitta@case.edu.
References
Author notes
H.H. and W.X. contributed equally.