Abstract

Introduction
Graft versus host disease (GVHD) is the main cause of morbi-mortality after allogeneic stem cell transplantation (allo-SCT). Despite considerable advances in our understanding of the pathophysiology, nowdays anticipation of GVHD is an unresolved matter. Several single-nucleotide polymorphisms (SNPs) in cytokine genes have shown to be associated with donor-recipient alloreactivity and, ultimately, with SCT outcome. In the present study, we propose a novel predictive model based on both clinical and genetic (SNP) variables applying an innovative estimation linear regression model, the least absolute shrinkage and selection operator (LASSO), in a large cohort of HLA-identical sibling donor allo-SCT.
Patients and Methods
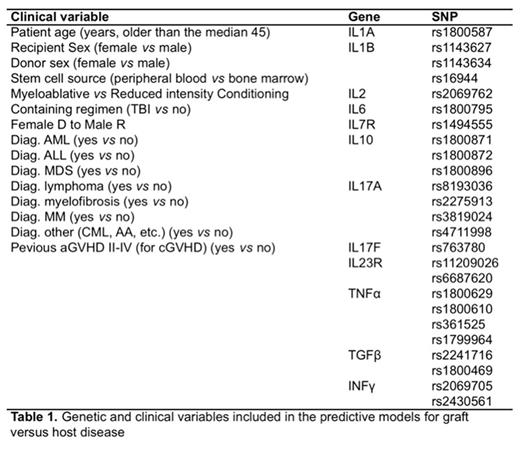
The study evaluated 25 SNPs in 12 genes (Table 1) in genomic DNA obtained from PB samples from 273 patients with available acute GVHD (aGVHD) data and 213 patients with chronic GVHD (cGVHD) data included in the DNA Bank of the Spanish Group for Hematopoietic Stem Cell Transplantation (GETH) and their HLA-identical sibling donors. Each SNP was assessed for different models of transmission (recessive, dominant, co-dominant and additive), producing 25 SNPs x 4 models = 100 variables. Clinical variables known to influence the development of GVHD were also considered (Table 1). Univariate regression analysis was performed using Cox regression (data not shown). Multivariant analysis was made with LASSO, an innovative estimation method for linear regression models which is able to select a set of optimal predictors from a large set of potential predictor variables and was considered as a variables selection method under the estimation of a Logit regression model. In this model, the strength of the penalty term is controlled by a smoothing parameter (λ), which is chosen by maximizing the area under ROC curve (AUC) and the correct classification rate (CCR). The statistical model was fitted (goodness-of-fit assessment) by randomly selecting the 85% of the data (the so-called "training set"), and the predictive ability was computed with the remaining 15% (the so-called "testing set"). In order to evaluate the performance and the prediction ability of each model, training and testing samples were randomly selected a total of 100 times. The distribution of the CCR and the AUC over the 100 samplings, were shown by means of box plots and statistical summary in the results data. Finally, for prediction purposes, we considered a cut-off value according to the proportion of Y=1 in the sample (0.28 for grades II-IV aGVHD, 0.11 for grades III-IV aGVHD and 0.30 for extensive cGVHD).
Results
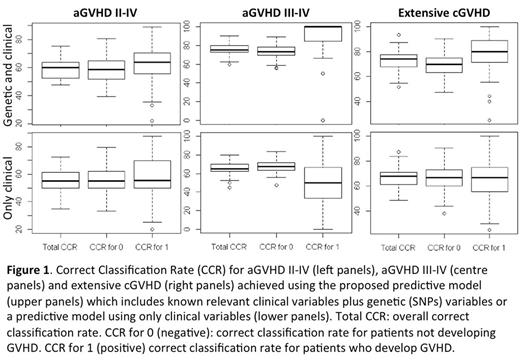
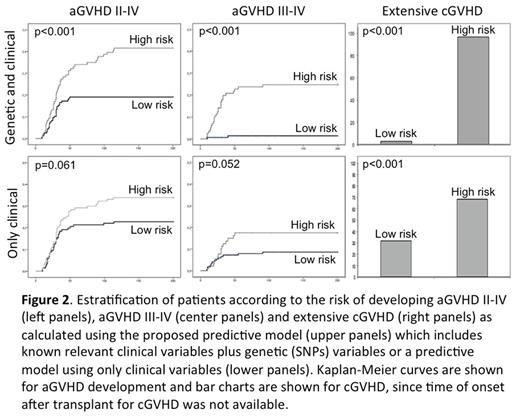
The best model to predict aGVHD II-IV included 11 genetic variables and no clinical variables with a CCR for patients who developed (CCR1) aGVHD II-IV of 63.6% (Figure 2). The best model to predict aGVHD III-IV included 20 genetic and 7 clinical variables with a CCR1 for aGVHD III-IV of 100%. The best model to predict extensive chronic GVHD included 10 genetic and 3 clinical variables with a CCR1 for extensive cGVHD of 80%. On the other hand, predictive models with only clinical variables showed a poorer CCR1 for patients who developed aGVHD II-IV, aGVHD III-IV and extensive cGVHD (55.6%, 50% and 66.7% respectively; Figure 1). Based on the results from LASSO multivariate analyses, a risk score was calculated for grades II-IV and III-IV aGVHD as well as for cGVHD and extensive cGVHD. Patients were categorized into two groups: low risk (below the cut-off value) and high risk (above the cut-off). Such risk model was able to stratify patients who develop grades II-IV aGVHD (p<0.001), grades III-IV aGVHD (p<0.001) and extensive cGVHD (p<0.001) more consistently than models only considering clinical variables (Figure 2).
Conclusions
Identification of biomarkers useful for the estimation of the risk of GVHD constitutes an unmet need in the clinical management of GVHD. The novel predictive model proposed here, based on clinical and genetic factors, allows significantly improved anticipation of aGVHD III-IV (100% accuracy) and extensive cGVHD (80%) after HLA-identical sibling donor allo-SCT. This approach would allow a personalized risk-adapted clinical management of patients after transplantation.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal