Abstract
Introduction:
A huge amount of data on genetic alterations has been compiled by high throughput sequencing studies in several hematologic malignancies. A molecular based categorization of these abnormalities could be helpful for further diagnostic and therapeutic approaches. Based on the type of alterations we discriminate 3 major categories: 1. copy number gains or losses, 2. fusion genes, and 3. molecular mutations. Studies on copy number data revealed that distinct tumor entities show typical but rather unspecific genetic alterations. Some fusion genes, e.g. involving FGFR, occur in diverse cancers, but others can also be specific for distinct diseases, like PML-RARA which is only found in APL. In particular kinase fusions, like BCR -ABL1 in CML or ALL, have tremendous clinical impact as affected patients respond to specific kinase inhibitors. However, there is limited knowledge yet on the entity-specificity for many novel gene mutations found in hematologic malignancies and a comprehensive analysis of their specificity is missing.
Aim:
Categorize gene mutations most frequently occurring in MDS and AML into entity-specific mutations, i.e. to distinguish MDS/AML-specific mutations from "pan-hematologic" or "pan-cancer" mutations.
Materials and Methods:
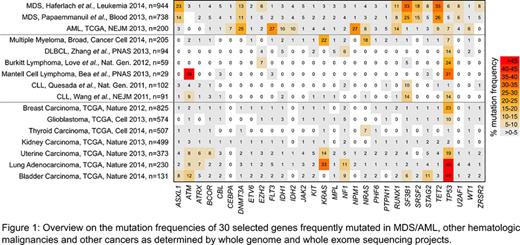
We i) selected the 65 most frequently mutated genes according to 3 major sequencing studies investigating the molecular basis of MDS and AML (n=1882 patients; Papaemmanuil et al., Blood 2013; Haferlach et al., Leukemia 2014; TCGA, NEJM 2013), ii) evaluated their mutation frequencies in whole genome/exome studies of other hematologic malignancies (including CLL, DLBCL, MCL, Burkitt lymphoma and MM, see Fig.1 for references) and solid tumors (as accessible via http://www.cbioportal.org, including glioblastoma, breast, lung, thyroid, kidney, uterine, and bladder carcinoma).
Characteristic for MDS as well as AML: Three genes were recurrently mutated in both entities but rarely in other hematologic malignancies and solid tumors: DNMT3A (MDS: 11-13%, AML: 25%, highest frequency (max) others: 5% in bladder), RUNX1 (MDS: 8-11%, AML: 14%, max others: 4% in bladder), TET2 (MDS: 26-33%, AML: 9%, max others: 8% in melanoma).
Characteristic for MDS was SRSF2, the only gene with a mutation frequency ≥5% in MDS but ≤2% in other entities (15-18% in MDS vs. 2% in bladder). Further, SF3B1 mutations were frequent in MDS (25-33%), CLL (10-14%), and solid tumors (max: 8%). ASXL1 mutations (14-23% in MDS) were also found in colorectal adenocarcinoma (10%) and ZRSR2 mutations (5-8% in MDS) in uterine cancer (3%).
Regarding AML, we identified 2 genes with a mutation frequency ≥5% in AML but ≤2% in any other entity: NPM1 (27%, max others: 2% in lung) and CEBPA (7%, max others: 2% in bladder). FLT3 mutations occurred significantly more often in AML (27%) than in other entities (max: 8% in melanoma; p<0.001). Other genes recurrently mutated in AML (≥5%) but rarely in other entities include IDH2 (10%, max others 8% in melanoma) and CBFB (6%, max others: 4% in breast). IDH1 mutations were more frequent in brain/glioma (76%) than in AML (10%).
Analyzing the mutated genes grouped into different biologic processes revealed MDS, Burkitt lymphoma, uterine and bladder cancer sharing the majority of affected processes, i.e. regulation of transcription, gene expression, splicing, chromatin organization and cell cycle. In contrast, the pattern was completely different in AML and involved cell proliferation, differentiation and several signaling pathways.
Conclusions: The majority of recurrently mutated genes in AML/MDS is rarely altered in other hematologic malignancies or solid tumors. While DNMT3A, RUNX1, and TET2 were linked to AML and MDS, we also found highly entity-specific genes either associated with AML or MDS. These can clearly be considered as driver genes and are of utmost diagnostic and/or therapeutic interest. Mutations of NPM1 and CEBPA turned out to be specific for AML supporting their currently discussed roles as distinct WHO entities. SRSF2 mutations were specifically associated with MDS making SRSF2 a prime candidate for further research on targeted therapies. Furthermore, the broad spectrum of mutated genes co-operatively impacting distinct biological processes in MDS, AML and the other analyzed malignancies supports its continued evaluation by comprehensive multi-gene screenings aiming at patient-specific and biology-based targeted therapies.
Rose:MLL Munich Leukemia Laboratory: Employment. Haferlach:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Kern:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Haferlach:MLL Munich Leukemia Laboratory: Employment, Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal