Abstract
Introduction: Activating mutation of FLT3 by internal tandem duplications (ITD) is the most common molecular aberration found in AML. FLT3-ITD mutation induces a proliferation signal and is associated with leukocytosis and poor prognosis. The co-existence of FLT3-ITD with translocation t(5:11) that yields fusion transcript of Nucleoporin98 and the nuclear receptor binding SET-domain protein 1 (NUP98/NSD1) was recently reported to be associated with chemo-resistance. Relapse risk of the majority of FLT3-ITD positive AML patients who are negative to NUP98/NSD1 is increased compared to other AML with normal karyotype, despite similar remission rates. The mechanism of FLT3-ITD contribution to relapse development in patients who achieved remission is unknown.
Aims: To explore potential mechanisms that allow relapse in FLT3-ITD positive AML in the presence or absence of NUP98/NSD1 fusion gene. In addition, time sequence of the emergence of both mutations is suggested from a case with relapse.
Methods: Leukemic blasts derived from newly diagnosed or relapsed FLT3-ITD positive AML patients were enriched for FLT3-ITD mutated sub-clones by sorting according to CD34 expression. Genomic DNA was extracted from each sorted fraction and FLT3-ITD mutation allele load was quantitatively determined by GeneScane. Total RNA was extracted from FLT3-ITD positive patients and RT-PCR was performed to detect NUP98/NSD1 fusion mRNA. All patients received induction with intensive chemotherapy combination of Daunorubicin and Cytarabine and their outcome was recorded. Chemo-sensitivity assays using Ara-C were conducted on different leukemia cell lines sub-populations divided by their CD34 expression.
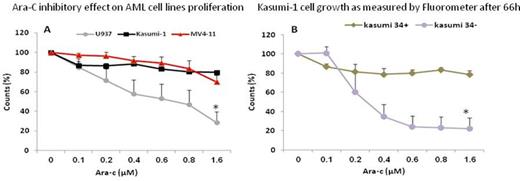
Results: NUP98/NSD1 was identified in 4 of 19 (21%) FLT3- ITD positive adult AML patients with normal karyotype who had a compatible donor and were considered transplant eligible. Patients harboring both NUP98/NSD1 and FLT3-ITD (75%) had higher rate of induction failure than FLT3-ITD patients without NUP98/NSD1 (40%). To explore the correlation between FLT3-ITD and differentiation capacity, primary AML FLT3-ITD positive blasts were sorted into two distinct sub-populations according to CD34 expression (example is shown in fig. 1). In 14/19 patients DNA from CD34+ and CD34- leukemic blasts was successfully extracted and FLT3- ITD allele load was tested and recorded as the ratio between FLT3 normal and mutated alleles. Of these 14 patients, 3 experienced induction failure and 8 (58%) achieved complete remission but unfortunately eventually relapsed. FLT3-ITD allelic ratio (AR) was equally measured in both CD34+ and CD34- sub-populations in 7 patients (50%) while in 5 patients (35.7%) the mutated allele was restricted to the CD34 positive cells and 4/5 (80%) patients experienced relapse. In two patients (14%) who also expressed NUP98/NSD1, the FLT3-ITD mutated allele was restricted to CD34 negative sub-population. In one patient, NUP98/NSD1 was detected in relapse but not in diagnostic specimen. Cytotoxic assay confirmed that differentiation stage as determined by CD34 expression is fundamental for chemo-sensitivity of leukemic cells regardless of their genetic profile. CD34 positive cell lines as well as CD34+ sub-population of Kasumi-1 cell line were resistant to the Ara-C compare to CD34 negative cell lines or the matched Kasumi-1 CD34- sub-population (fig. 2).
Conclusions: FLT3-IDT mutation is a late event during leukemogenesis. We observed NUP98/NSD1 that emerged as a new mutation on relapse in a FLT3-IDT positive patient, assuming this may also be a later event. Our strategy to sort blasts according to their CD34 expression enable us to describe the accumulation of FLT3-ITD sub-clones at the primitive stage that express CD34, therefore, these leukemic sub-clones may be enriched with early leukemic precursors. Such differentiating blockage results a higher portion of cells which survive chemotherapy among FLT3-ITD and hence increases relapse risk as an indirect effect. Inducing differentiation in FLT3-ITD positive AML should be further studied as a therapeutic strategy.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal