Abstract
Background: Recent years, the incidence and mortality of fungal infection has been on the rise in the patients with hematologic malignancies. This is mainly associated with antifungal resistance and the restricted number of available antifungal drugs. Candida species is one of the most prevalent pathogens in these immunodeficient patients. However, the study of azole resistance mechanisms of Candida has focused on C.albicans, C.glabrata, C.tropicalis. And few studies talked about resistance mechanisms of C.krusei, especially resistant to itraconazole. It was reported that the mutation or overexpression of 14¦Á-demethylases (encoded by ERG11) and upregulation of efflux transporters (encoded by ABC1 and ABC2) may be involved in azole resistance of C.krusei. Here, The purpose of the present study is to preliminarily explore the main molecular mechanisms responsible for Candida krusei clinical isolates to itraconazole, and may provide new sight into fungal infection therapy.
Methods: The 14¦Á-demethylases encoded by ERG11 gene in the 16 C.krusei clinical isolates were amplified by polymerase chain reaction (PCR), and their nucleotide sequences were determined to detect point mutations. Meanwhile, ERG11 and efflux transporters (ABC1 and ABC2) genes were determined by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) for their expression in itraconazole-resistant (R), itraconazole-susceptible dose dependent (SDD) and itraconazole- susceptible (S) C.krusei at the mRNA level.
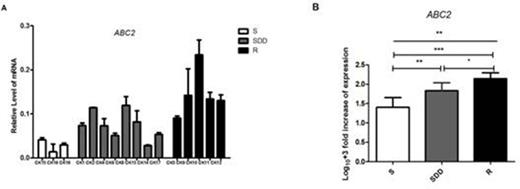
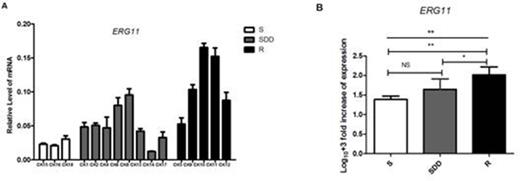
Results: We found 7-point mutations in ERG11 gene of all the C.krusei clinical isolates, including 6 synonymous mutations and 1 missense mutation (C44T). However, the missense mutation was found in the three groups. The mRNA levels of ERG11 gene in itraconazole-resistant isolates showed higher expression compared with itraconazole-susceptible dose dependent and itraconazole- susceptible ones (P=0.015 and P=0.002 respectively). ABC2 gene mRNA levels in itraconazole-resistant group was significantly higher than the other two groups, and the levels of their expression in the isolates appeared to increase with the decrease of susceptibility to itraconazole (P=0.007 in SDD compared with S, P=0.016 in SDD with R, and P<0.001 in S with R respectively). While ABC1 gene presented lower expression in itraconazole resistant strains. However, the mRNA levels of ERG11, ABC1 and ABC2 in a C.krusei (CK10) resistant to both itraconazole and voriconazole were expressed highest in all the itraconazole-resistant isolates. The relative mRNA levels of gene ABC2 and ERG11 can be found in Fig.1 and Fig.2 respectively.
Conclusions: There are ERG11 gene polymorphisms in clinical isolates of C.krusei. ERG11 gene mutations were not found to be involved in the development of itraconazole resistance in C.krusei. ERG11 and ABC2 overexpression might be responsible for the acquired itraconazole resistance of these clinical isolates. Therefore, combination of azole and selective efflux transporter inhibitors may help reverse azole resistance and enhance antifungal effect.
ABC2 relative gene expression levels in three groups of C.krusei clinical isolates. (A) Relative levels of ABC2 mRNA in all the C.krusei clinical isolates. ABC2 gene expression levels was quantified and normalized relative to the housekeeping gene, ACT1; S, itraconazole-susceptible; SDD, itraconazole-susceptibledose dependent; R, itraconazole-resistant. (B) Log10+3 fold increase of gene expression levels in three groups. (*P<0.05 in R compared with SDD; **P<0.01 in SDD with S; ***P<0.001 in R with S)
ABC2 relative gene expression levels in three groups of C.krusei clinical isolates. (A) Relative levels of ABC2 mRNA in all the C.krusei clinical isolates. ABC2 gene expression levels was quantified and normalized relative to the housekeeping gene, ACT1; S, itraconazole-susceptible; SDD, itraconazole-susceptibledose dependent; R, itraconazole-resistant. (B) Log10+3 fold increase of gene expression levels in three groups. (*P<0.05 in R compared with SDD; **P<0.01 in SDD with S; ***P<0.001 in R with S)
ERG11 relative gene expression levels in three groups of C.krusei clinical isolates. (A) Relative levels of ERG11 mRNA in all the C.krusei clinical isolates. ERG11 gene expression levels was quantified and normalized relative to the housekeeping gene, ACT1; S, itraconazole-susceptible; SDD, itraconazole-susceptibledose dependent; R, itraconazole-resistant. (B) Log10+3 fold increase of gene expression levels in three groups. (NS, no significance in SDD compared with S; *P<0.05 in R with SDD; **P<0.01 in R with S)
ERG11 relative gene expression levels in three groups of C.krusei clinical isolates. (A) Relative levels of ERG11 mRNA in all the C.krusei clinical isolates. ERG11 gene expression levels was quantified and normalized relative to the housekeeping gene, ACT1; S, itraconazole-susceptible; SDD, itraconazole-susceptibledose dependent; R, itraconazole-resistant. (B) Log10+3 fold increase of gene expression levels in three groups. (NS, no significance in SDD compared with S; *P<0.05 in R with SDD; **P<0.01 in R with S)
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal