Abstract
Background: Regulatory CD4 T cells (Treg) are potent to suppress the responses of conventional CD4+ (Tcon) and CD8+ T cells to alloantigens and prevent graft-vs.-host diseases (GVHD). Since the mechanisms for thymic selection of Treg and Tcon are distinct, we hypothesized that the two cells types use distinct TCR repertoires in the response to same alloantigen. We have used high throughput deep sequencing to study the T cell receptor (TCR) Vbeta CDR3 repertoire of CD4 Treg and CD4 Tcon at baseline and again after expansion with alloantigen.
Methods: CD4+CD25+CD127- Treg and CD4+CD25-CD127- Tcon were FACS-sorted from healthy donor PBMCs or umblical cord blood mononuclear cells. Aliquots of sorted Treg and Tcon were expanded separately by DC from the same Major Histocompatibility (MHC)-mismatched donor. Treg were cultured with DCs, IL-2, IL-15 and rapamycin, while Tcon were cultured with DCs and IL-2. Genomic DNA from baseline or expanded purified Treg and Tcon was sequenced, at least 10^6 deep, by Adaptive Biotechnologies multiplex kit to detect unique T cell receptor (TCR) Vbeta CDR3 sequences. Data was analyzed by the algorithm established for the Adaptive Biotechnologies software and characterized according to the IMGT (International ImmunoGeneTics information system) nomenclature.
Results: The TCR Vbeta CDR3 sequences of baseline natural Treg and Tcon T cell have minimal overlap, indicating that the thymus shapes distinct TCR repertoires in these two cell types. However, the CDR3 length and the frequency of nucleotide deletion or insertion at the Vbeta-Dbeta and Dbeta-Jbeta junction were similar. By employing the "robust regression model", we identified expanded TCR Vbeta CDR3 sequences among both Treg and Tcon after in vitro culture with the same alloantigens. These expanded Treg and Tcon used unique TCR Vbeta CDR3 sequences that are not shared by the other cell type. Expanded Treg and Tcon displayed fewer nucleotide deletions and insertions at the Vbeta-Dbeta and Dbeta-Jbeta junction than at baseline. The frequency of the expanded sequences, insertions and deletions were of the same magnitude in Treg and Tcon suggesting that both undergo similar processes of antigen-driven TCR selection and magnitude of cell expansion in vitro.
Conclusion: CD4 Treg and Tcon utilize largely distinct TCR Vbeta CDR3 repertoires at baseline and after expansion against the same alloantigens. Questions remain whether Treg and Tcon recognize the same or distinct peptides from the same antigen, and whether they bind peptide with different avidity. TCR Vbeta CDR3 deep sequencing ought to be used to track single Treg and Tcon after adoptive cell therapy.
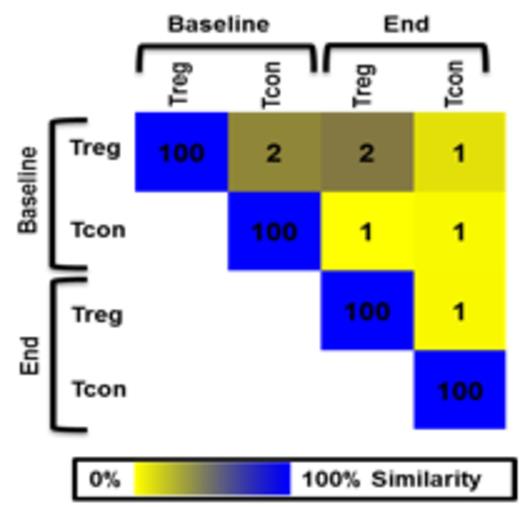
CDR3 nucleotide sequences of natural Treg and Tcon do not overlap before (Baseline) and after (End) expansion against alloantigen
CDR3 nucleotide sequences of natural Treg and Tcon do not overlap before (Baseline) and after (End) expansion against alloantigen
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal