Abstract

Introduction
Multiple myeloma (MM) is characterized by a highly variable disease course, which can be traced to initiating and acquired genomic events. Whole exome analysis of matched tumor and germline DNA from 287 MM patients identified recurrently somatically mutated genes (RSMGs) (Lohr et al. - Cancer Cell 2014, Bolli et al. - Nat Commun 2014). Despite the fact that these RSMGs affect pathways that are biologically important in MM, the clinical relevance of many of these genes in the context of conventional prognostic markers remains to be elucidated.
Aims
The aims of this pilot study were: (1) To validate the prevalence of RSMGs in our newly diagnosed MM patient cohort; (2) To assess the correlation between RSMGs, clinical parameters and outcome; (3) To thereby identify the potential clinical usefulness of introducing RSMG mutational profiling in larger MM trial cohorts.
Material and Methods
CD138+ enriched MM cells and peripheral blood were obtained with informed consent from chemotherapy-naive patients, participating in 3 clinical trials: HOVON-65/GMMG-HD4, HOVON-87/NMSG-18 and Carthadex (EudraCT number 2004-000944-26, 2007-004007-34 and 2009-014922-40, respectively).
Matched tumor and germline DNA were sequenced on an Ion Torrent sequencing platform (PGM, Life Technologies), using the M3 P Mutational Panel v3.0, comprising 1327 customized oligos (Life Technologies), targeted at the coding sequences of 88 MM-relevant genes, including the RSMGs. Somatic mutations were considered positive when present in >=10% of tumor reads and <=10% germline reads, with a minimal coverage of 10x and being non-synonymous, or splice donor variants.
All statistical analyses were performed in SPSS version 23, using the log-rank and Mann-Whitney U-test, with the Bonferroni test to correct for multiple comparisons.
Results
A total of 206 DNA samples were sequenced from 103 patients (HOVON-65/GMMG-HD4 (n=16), HOVON-87/NMSG-18 (n=67), Carthadex (n=20)) with an average coverage of 574x in tumor DNA, 451x in germline DNA and an overall coverage of 98%. We collected follow-up data from 102/103 patients, with a median follow-up time of 30 months.
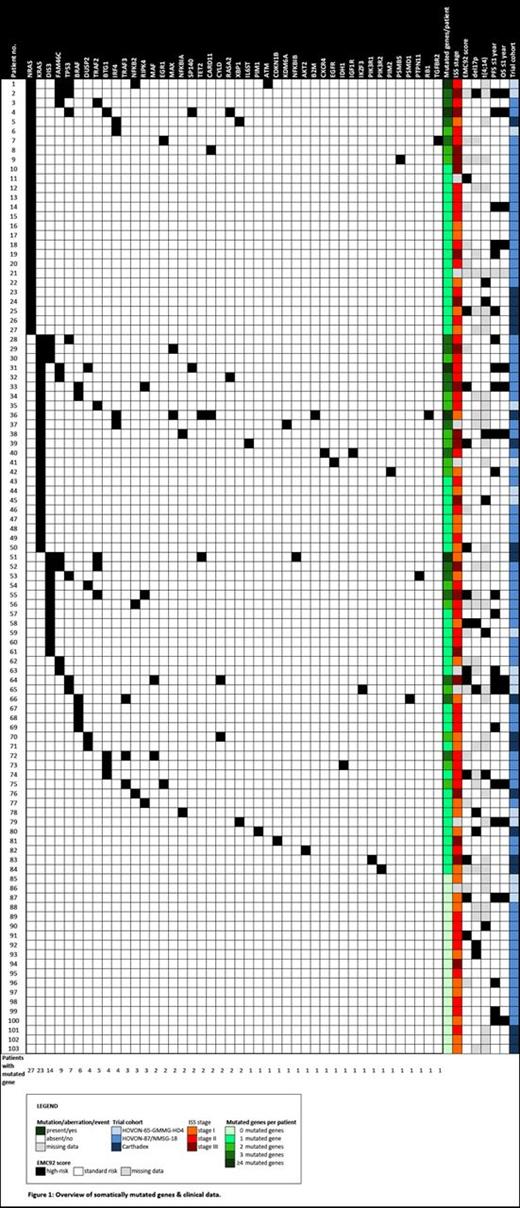
168 somatic mutations were detected in 44/88 genes. 82% of patients had at least 1 somatic mutation. Genes most frequently mutated were: (1) NRAS (26%), (2) KRAS (22%), (3) DIS3 (14%), (4) FAM46C (9%), (5) TP53 (7%) and (6) BRAF (6%) (Figure 1). Of note, NRAS and KRAS mutations were mutually exclusive in our cohort. Moreover, all TP53 mutations were located in its DNA binding domain. Three out of 6 BRAF mutations were predicted to cause a V600E amino acid change.
We focused on these 6 RSMGs in all further analyses. Correlating mutational status with Progression Free Survival (PFS) and Overall Survival (OS) showed that TP53 mutated patients had a significantly shorter PFS compared to those with wildtype TP53 (adj. p-value=0,018; n=7 versus n=95). Comparing the mutational status of the 6 RSMGs, transplant versus non-transplant protocol, number of mutated genes in the M3 P panel, del17p and t(4;14) status, EMC92 score and ISS stage between patients with a PFS <=1 year and >1 year (n=23 versus n=79), only showed a significant correlation with TP53 mutational status (adj. p-value=0,012). TP53 mutational status remained the only significant prognostic factor when comparing patients with an OS <=1 year and >1 year (adj. p-value=0,003; n=13 versus n=89).
When comparing the number of mutated genes, del17p and t(4;14) status, EMC92 score, transplant versus non-transplant protocol and ISS stage between TP53 mutated and wildtype MM, TP53 mutated patients had a significantly higher number of mutated genes in the M3 P panel (adj. p-value=0,001).
Conclusions
(1) With the M3 P Mutational Panel, we confirm the published prevalence of RSMGs in MM in our cohort of chemotherapy-naive patients. NRAS, KRAS, DIS3, FAM46C, TP53 and BRAF are the most frequently mutated genes.
(2) TP53 mutational status is the strongest unfavorable prognostic factor in our cohort and it seems to be associated with greater mutational burden. Validation in a more extensive population is planned.
(3) This warrants further investigation of the mutational status of these genes in larger clinical trial cohorts, enabling a more robust comparison with conventional prognostic markers in a multivariate analysis.
Broijl:Celgene: Membership on an entity's Board of Directors or advisory committees; Amgen: Membership on an entity's Board of Directors or advisory committees. Zweegman:Takeda: Research Funding; Janssen: Research Funding; Celgene: Research Funding. Sonneveld:Janssen: Honoraria, Research Funding; Celgene: Honoraria, Research Funding; Amgen: Honoraria, Research Funding; Karyopharm: Research Funding; SkylineDx: Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal