Abstract
Background
Familial form of acute myeloid leukemia (AML) is not well known except for extreme cases. BRCA1/2 and p53 germline mutation are well known genetic changes that is related to familial AML¡¯s. Because of diverse penetrance potential of germline mutations and complex pathogenesis for the development of AML, it is not easy to discern familial form of AML. In this study, we performed next generation sequencing (NGS) study of a family who has been suspected to have familial form of AML.
Material and method
Two relatively young patients have been referred to our institution for AML. One was a 41 years old male (son) and the other was 58 years old female (mother). To discover genetic factors involved in hematological familial cancer syndrome, we performed WGS/WTS using DNA of mother and son who are AML patients and WGS using DNA of healthy father. Tumor DNA and RNA of mother and son were extracted from bone marrow samples which were collected at the time of diagnosis and control DNA and RNA was extracted from saliva and bone marrow samples, respectively, at the time of CR. WGS were performed using HiSeq X ten (illumina, San Diego, USA). For WTS, we used HiSeq2000 platform (illumina, San Diego, USA). For the analysis of WGS, we used GATK unifiedGenotyper for caller in this study. The first filter was set an average depth >10 and conf_cut > 50. And next filter step was using several databases (dbSNP, clinvar, cosmnic70, nci60) and filtering tools (Meta SVM and Meta LR) to filter out of called loci. Meerkat was used to analyze structure variations (SVs). To analyze DEGs using WTS, we used HTSeq-count. To analyze DEGs, we used WTS and HTSeq-count.
Results and discussion
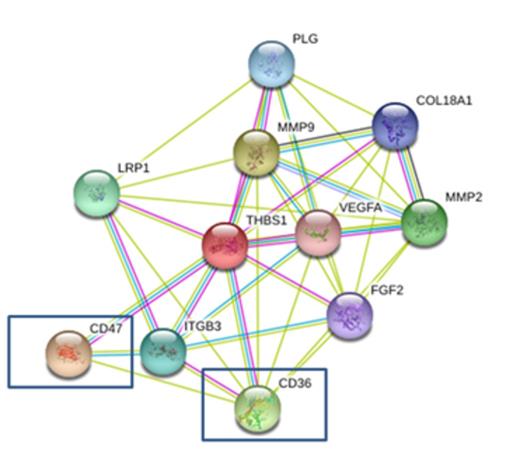
A total of 3,695,266 loci of normal DNA and 3,977,321 loci of tumor DNA were found in mom samples and 3,622,083 of these were commonly found in both normal and tumor samples. Similarly, a total of 3,513,806 loci of normal and 3,935,873 loci of tumor DNA were found in son samples, of which 3,476,405 were commonly found. Of these common loci, we identified 2,799,429 that are universally present in both mum and son. To determine genes that are found only in patients (Mon and son), we filtered out the loci of father¡¯s SNPs and excluded 2,191,882 loci We subsequently identified a total of 607,547 candidate loci which have association with AML malignancies. Using Support vector machines (SVMs) and DNM filter, we found 37 significant genes that are considered to be related with de-novo AML Among these, 12 were (AGL, COL12A1, IMPDH1, LIPN, MET, MYH13, PBX3, ROBO3, SLC34A3, SMO, THBS1 and TP63) already reported to have an association with hematopoietic disorder, while 25 were a novel mutation genes. The most interesting gene is THBS1. It is reported to affect hematopoietic differentiation function via CD36 and CD47 and a greater than 3 fold change was found in both mum and son in DEG analysis. In DEG analysis, total 1317 gene in mother and 473 genes in son were shown differently expressed above 3FC. The number of recurrent DEGs between mother and son is 144 and these genes were estimated to involve in Systemic lupus erythematosus, Chemokine signaling and bladder cancer pathway.
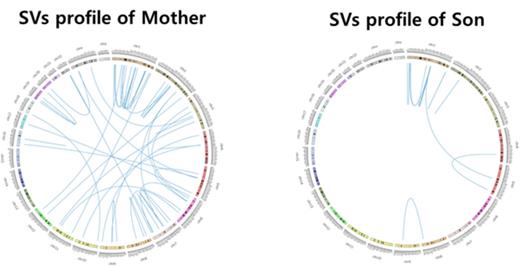
We used Mutect to detect somatic mutation that acts as second hit. 29 nonsynonymous-SNVs were detected in son sample and 43 nonsynonymous-SNVs were detected in mother sample. OR11H1 gene was recurrently shown in both mother and son. We performed Structure variants analysis to identify second hit SVs using Meerkat in mother and son separately SVs of mother were detected in several regions and 69 genes were detected in exonic regions, 39 genes were somatic SVs and 12 of these were filtered out because the genes were also detected in the normal sample of son. 27 SVs genes are considered as a candidate of AML second hit in mother. On the other hand, we found 5 genes somatic SVs in son using the same method. Of note, FAM231A gene overlapped with mom tumor sample and son tumor sample.
In conclusion, we found 37 significant genes may be related with de-novo AML. In addition, several genetic factors affect tumorigenesis through second hit.
Association between THBS1 and other genes by gene to gene networking
Association between THBS1 and other genes by gene to gene networking
Expression level and heatmap of gene expression of mother and son by DEGs analysis
Expression level and heatmap of gene expression of mother and son by DEGs analysis
Circosplot of Structure Variantions(SVs) of mother and son
Circosplot of Structure Variantions(SVs) of mother and son
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal