Abstract
Introduction:
Cytogenetic analysis is well integrated into the work up of acute myeloid leukemia (AML) and myelodysplastic syndromes (MDS). In fact, cytogenetics constitute the single most important prognostic indicator for AML and are frequently found in MDS (50-80%). It may be assumed that their prognostic impact is due to loss of candidate genes that lie in abnormal regions [e.g., MEGF1 in del(5q)]. The advantage of karyotype or fluorescence in-situ hybridization (FISH) testing is that they offer a high level of view of the genome detecting large structural changes that may not be amenable to evaluation by next-generation sequencing (NGS). Conversely, the resolution of karyotype is insufficient to detect single nucleotide variants better evaluated by NGS. Thus, each offers unique and complementary genetic data. In this study we investigated associations between commonly mutated genes in myeloid disease by NGS and cytogenetic abnormalities (CA) detected by karyotyping/FISH studies in the hopes of uncovering possible cooperative mechanisms of disease.
Materials and Methods:
All patients between May 2011 and October 2014 with hematopoietic malignancies and available mutational analysis by NGS were included in this study. All NGS was done in a CAP/CLIA certified laboratory environment, using a 5 gene panel initially and later a 21 gene panel. A subset of cases were evaluated on 405 gene panel. Karyotyping was performed at a CAP/CLIA certified lab using standard procedures. FISH studies were performed at our center.
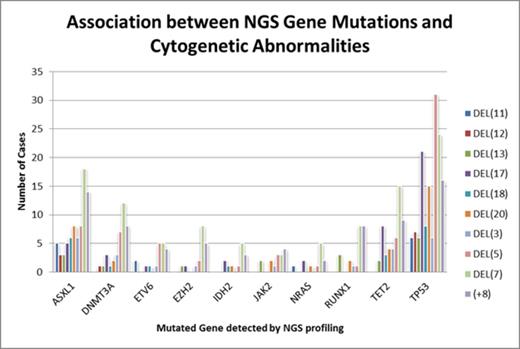
Results:
Four hundred and ninety patients were tested for mutation by the 5 or 21 gene panel, while 33 patients were tested by a 436 gene panel (total n=523). There were 186 (35.6%) AML, 165 (31.5%) MDS, 44 (8.4%) MPN, 44 (8.4%) MDS/MPN, 42 (8%) others (including aplastic anemia, lymphoma, ALL, and multiple myeloma), and 42 (8%) cases with normal morphology. Of these patients, 267 (51%) had cytogenetic abnormalities (CA) and 358 (69%) patients had gene mutations. The data for most common mutations and CA are summarized in Figure 1. The most common CA were: del(7) 24%, del(5) 20.6%, +8 15%, del(20) 12%, and del(17) 8.8%. From all genes interrogated, 80 genes were found to be mutated. TET2 had the highest mutation rate (19.2%), followed by ASXL1 (17.9%) and DNMT3A (13%). TP53 had a mutation rate of 11.1%. However, TP53 mutated cases were most likely to harbor a concurrent CA (85% of TP53 mutated patients). Other frequently mutated genes, namely TET2, ASXL1, DNMT3A, JAK2, RUNX1, had a concurrent CA nearly half of the time (42.1%, 46.5%, 43.9%, 34.2%, 47.8%, respectively). The most frequent mutation and CA combinations were TP53 with del(5) (8%), TP53 with del(7) (6.2%), TP53 with del(17) (5.5%), and TP53 with trisomy 8 (4.2%). TET2 and ASXL1 tended to co-occur with del(7) (4% each). There appears to be a preferential association between TP53 mutation and chromosomal deletions. As previously known, FLT3 and NPM1 mutations were more likely to occur in patients without CA. ASXL1 mutation and del(7) co-occurred in 18 of the patients, 15 of which are AML patients (83%). TP53 and del(5), which is the most frequent combination, occurred in 31 patients, 14 (45.2%) of which were AML patients. Similarly, 52.4% of patients with TP53 mutations and del(17), and 50% of patients with TP53 mutations and +8 were AML patients. Therefore, the percentages of gene mutations co-occurring with CA were higher in AML than in other categories of disease.
Conclusions:
TET2 and ASXL1 are the most common mutations in the hematologic malignancies tested, but TP53 mutations are most likely to coincide with concurrent CA, while FLT3 and NPM1 are less likely to do so. Del(5) is the CA most commonly found in cases with gene mutations detected by NGS. Concurrent gene mutations and CA [e.g., ASXL1 with del(7), TP53 with del(17), TP53 with trisomy 8, TP53 with del(5)] are enriched in AML patients indicating possible cooperation of mutation and genes housed within these structural abnormalities in the pathogenesis of acute myeloid leukemia.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal