In this issue of Blood, Van der Meulen et al demonstrate that the lysine demethylase UTX (also known as KDM6A), best known as an “eraser” of methylation at histone 3 lysine 27 (H3K27), is an X-linked tumor suppressor in T-cell acute lymphoblastic leukemia (T-ALL).1 This finding is surprising because the biochemical activity of UTX at H3K27 is antagonized by the “writer” methyltransferase EZH2, which also functions as a tumor suppressor in this disease.
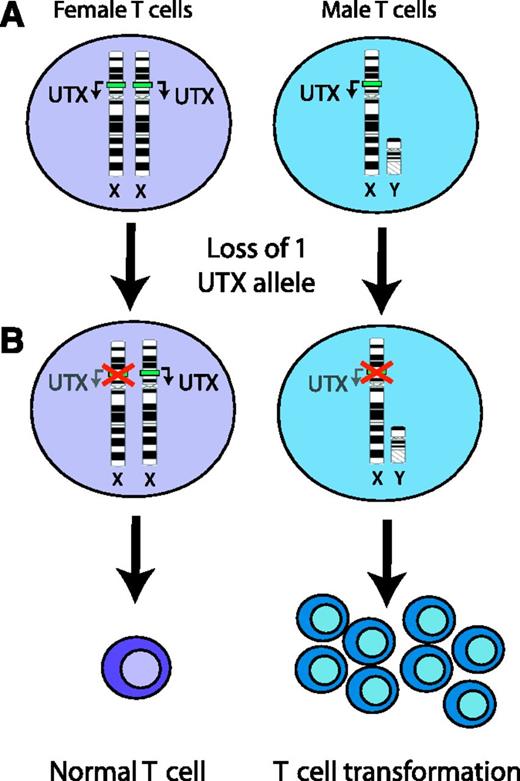
UTX is an X-linked tumor suppressor in T-cell leukemogenesis. (A) The UTX demethylase is within the nonautosomal region of chromosome X. Importantly, expression of this gene escapes X inactivation, and both alleles are expressed in female cells. (B) Following mutational inactivation of 1 UTX allele, female cells continue to express the wild-type allele, which appears to be sufficient for tumor suppression. By contrast, loss of the only UTX allele in male cells results in complete inactivation of this tumor suppressor, which promotes T-cell transformation.
UTX is an X-linked tumor suppressor in T-cell leukemogenesis. (A) The UTX demethylase is within the nonautosomal region of chromosome X. Importantly, expression of this gene escapes X inactivation, and both alleles are expressed in female cells. (B) Following mutational inactivation of 1 UTX allele, female cells continue to express the wild-type allele, which appears to be sufficient for tumor suppression. By contrast, loss of the only UTX allele in male cells results in complete inactivation of this tumor suppressor, which promotes T-cell transformation.
T-ALL is an aggressive malignancy with a higher incidence in males, yet the biologic basis for this clinical observation remains poorly understood. One potential explanation for gender bias is the presence of tumor suppressor genes on the nonautosomal region of chromosome X. Females harbor 2 copies of such genes whereas males have only 1. A mutation of a single allele would thus be sufficient to completely inactivate such a gene in male but not in female cells. However, gender differences in gene copy number are balanced by X inactivation, which silences expression of most genes on an X chromosome in female cells. Recent work has identified candidate T-ALL tumor suppressors on the X chromosome, including PHF6 and RPL10.2,3 However, these genes both appear to undergo X inactivation in females, suggesting that these may not contribute to gender bias.2,3 Nevertheless, a subset of genes on the nonautosomal region of chromosome X escape X inactivation in females, and tumor suppressors among these might contribute to the male predominance of T-ALL. In this issue, Van der Meulen and colleagues report the identification of UTX as one such tumor suppressor that escapes X inactivation and is preferentially mutated in male patients with T-ALL (see figure).
The authors began by sequencing a cohort of primary T-ALL patient samples, which revealed mutations of UTX in 14% of 35 cases analyzed. All mutations identified were truncating, and clustered strikingly within the N-terminal region of the catalytic JmjC domain of UTX. To test the functional consequences of UTX inactivation, the authors first used short hairpin RNA (shRNA) knockdown in a murine T-ALL cell line whose viability is dependent on exogenously provided interleukin growth factors. UTX knockdown improved cellular fitness following growth factor deprivation, indicating that UTX inactivation can substitute for growth factor signaling in T-ALL cells.
To test the role of UTX in vivo, the authors then turned to a murine model of T-ALL based on transduction of hematopoietic cells with constitutively active NOTCH1, followed by transplantation into irradiated recipients, which reliably induces T-ALL. UTX knockdown significantly accelerated tumor onset in this model, indicating that UTX is a bona fide T-ALL tumor suppressor. These data are reinforced by a recent study demonstrating similar results in UTX knockout mice.4 Van der Meulen and colleagues then used gene expression profiling of T-ALL cells infected with control or UTX shRNA to assess the transcriptional consequences of UTX inactivation. This analysis revealed several candidate mediators of tumor suppression downstream of UTX inactivation, including transcriptional downregulation of the NF1 tumor suppressor and upregulation of the TAL1 oncogene in these cells.
UTX is a lysine demethylase best known as an “eraser” of H3K27 methylation. In cells, DNA exists in association with histone proteins and other molecules in a complex known as chromatin. Enzymatic modifications of histone proteins have been implicated in the regulation of gene expression. The H3K27 methylation mark is associated with chromatin condensation and transcriptional repression. The biochemical function of UTX on H3K27 is antagonized by methyltransferase enzymes, including EZH1 and EZH2. Thus, Van der Meulen and colleagues hypothesized that UTX-deficient cells might be more sensitive to inhibitors of these methyltransferases. Indeed, the authors found that UTX knockdown rendered leukemic cells more sensitive to DZNep, a small molecule whose cellular effects include inhibition of EZH2.
Intriguingly, although UTX and EZH2 have antagonistic methylation activity at H3K27, both UTX and EZH2 function as tumor suppressors in T-ALL.1,4-6 One potential explanation for this apparent discrepancy is that UTX and EZH2 may have distinct transcriptional targets. Indeed, additional H3K27 “writer” methyltransferases and “eraser” demethylases exist, so that UTX and EZH2 need not necessarily function as an antagonistic pair for the maintenance of balanced H3K27 methylation at individual loci. In fact, recent work has shown that JMJD3 (also known as KDM6B), a UTX-related H3K27 demethylase, is bound by NOTCH1 and required for the maintenance of NOTCH1-induced T-ALL.4 By contrast, UTX does not bind NOTCH1 and suppresses NOTCH1-induced tumorigenesis.4 Thus, a speculative model to explain these observations is that EZH2 and JMJD3 may function as an antagonistic “writer-eraser” pair, with the UTX “eraser” being antagonistic to a different H3K27 methyltransferase. Such a model would mimic the antagonistic relationships that are conserved across substrates for some kinase-phosphatase pairs.7
The tumor suppressor function of UTX could also be mediated by functions independent of its ability to demethylate H3K27. Such possibilities include demethylation of lysine residues on nonhistone substrates or demethylase-independent functions. For example, UTX has been shown to interact with the switch/sucrose nonfermentable (SWI/SNF) chromatin-remodeling complex,8 a key tumor suppressor in diverse human cancers. Indeed, recent data have demonstrated that the tumor suppressor BCL11B, which is recurrently inactivated in human T-ALL, is a dedicated SWI/SNF subunit.9,10 Thus, UTX deficiency might represent an alternative mechanism for disabling the tumor suppressor function of SWI/SNF in T-ALL.
Unraveling the role of chromatin-modifying enzymes in T-ALL has important potential therapeutic implications. The overexpression of oncogenic transcription factors plays key roles in T-ALL, yet transcription factors have proven to be largely intractable targets for drug development. However, transformation by oncogenic transcription factors is strongly influenced by the accessibility of their binding sites within DNA, which is highly regulated by chromatin-modifying enzymes such as UTX and EZH2. These enzymes are much more tractable therapeutic targets than transcription factors. Thus, unraveling the precise role of such chromatin-modifying enzymes in T-ALL pathobiology could reveal therapeutic strategies to specifically reverse oncogenic gene expression programs. Indeed, the work of Van der Meulen et al1 and of Ntziachristos et al4 provides a compelling rationale for testing inhibitors of 2 such enzymes, EZH2 and JMJD3, in molecularly defined subsets of T-ALL.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal