Abstract
Introduction
Acute myeloid leukemia (AML) is a highly heterogeneous disease with various cytogenetic and molecular abnormalities, some of which bear prognostic significance. Expression levels of some single microRNAs are influential for prognosis, but a system integrating several together and considering the weight of each should be more powerful. We sought to define a simple microRNA signature to predict prognosis in the patients.
Method
A cohort of 195 de novo AML patients who had cryopreserved marrow cells for study were subjected to global microRNA analysis. Analysis for overall survival (OS) was based on the 138 patients who received standard intensive chemotherapy (NTUH cohort). The AML cohort from the Cancer Genome Atlas (TCGA), which contains publically available data of microRNA and clinical information, serves as a validation cohort.
To build a risk scoring system based on the global microRNA expression, we first analyzed the association between OS and the expression levels of individual microRNAs using univariate Cox proportional hazards regression model. MicroRNAs whose expression levels harbored top significance on OS (univariate Cox P < 0.005) were then analyzed with the multivariate Cox model. These microRNAs with independent survival significance (multivariate Cox P < 0.1) were selected to generate a risk scoring system, in which the expression of component microRNAs went through another round of multivariate Cox regression test to get beta values as weights for each microRNA. We also analyzed the global mRNA expression profiles from ours and TCGA cohort to sort out the physiologic pathways associated with high/low scores.
Results
Eleven microRNAs were significantly associated with OS by univariate Cox analysis (P < 0.005). After introducing the expression of these microRNAs into a multivariate Cox model, we identified high expression of hsa-miR-9-5p and hsa-miR-155-5p were independently associated with poor OS, while that of hsa-miR-203 had a trend of association with favorable OS. We constructed a risk scoring system: Risk = 0.4908 [Zhsa-miR-9-5p] + 0.2243 [Zhsa-miR-155-5p] - 0.7187 [Zhsa-miR-203], where the weights of microRNAs are beta values from multivariate Cox analysis and ZmicroRNA means the levels of specific microRNA expression after z-transformation.
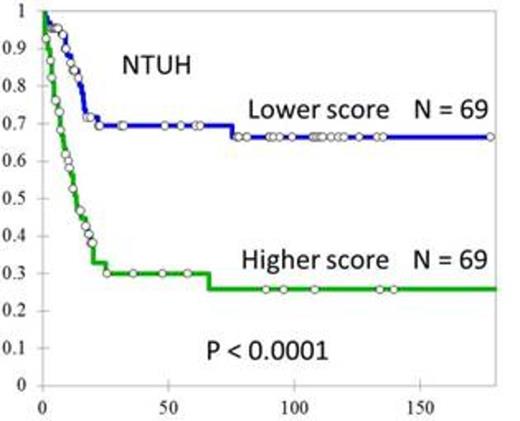
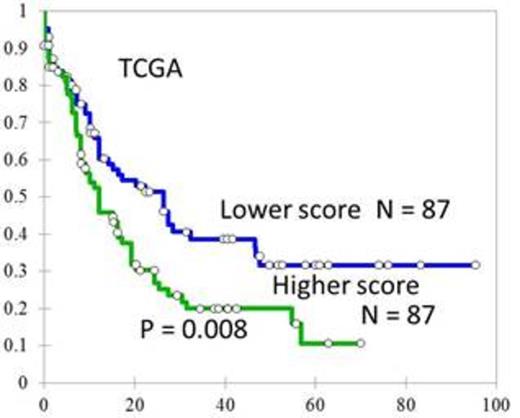
AML patients with higher scores had significantly shorter OS compared with those with lower scores in ours (median 13.5 months vs. not reached, P < 0.0001, Figure 1) as well as in the validation TCGA AML cohort (median 12.2 vs 26.4 months, P = 0.008, Figure 2). When restricting the analysis in our patients with a normal karyotype, the OS of patients with higher scores still fared worse (median 17.0 months vs. not reached, P = 0.006). Moreover, high score appeared to be an unfavorable prognostic factor independent of age, white cell count, cytogenetics, and gene mutation in both ours (HR=2.079, 95% CI 1.407-3.073, p<0.001) and TCGA cohort (HR=1.544, 95% CI 1.229-1.940, p<0.001). We also noted that the 3-microRNA signature outperformed all the single microRNA expression in prognostic significance. By analyzing the mRNA expression profiles, we sorted out several cancer-related pathways highly correlated with the microRNA prognostic signature. Since the scoring system was designed by z-transformed microRNA expression levels as inputs, cohort mean and cohort standard deviation of each of the three microRNAs are required for that formula. We provide the calculating formula we used in the NTUH dataset: Risk = 0.4908 (-Delta Cthsa-miR-9-5p+15.71)/3.60 + 0.2243 (-Delta Cthsa-miR-155-5p+6.94)/1.45 - 0.7187 (-Delta Cthsa-miR-203+17.16)/2.66. Here the Delta Ct values are Ct of the microRNA subtract Ct of the endogenous control MAMMU6; 15.71 and 3.60 are the mean and standard deviation of Delta Ct hsa-miR-9-5p. The same annotation applies to hsa-miR-155-5p and hsa-miR-203. For each newly diagnosed patient, the expression levels of hsa-miR-9-5p, hsa-miR-155-5p, hsa-miR-203, and MAMMU6 as determined by real-time PCR is sufficient to get a prognostic score, which will then be compared with our cohort median score 0.0031 to stratify the risk group.
Conclusion
We present a simple and user-friendly 3-microRNA signature as a powerful prognostic factor for AML through multiple rounds of statistical analyses on our cohort and further validation by another independent patient group.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal