Abstract
Introduction : Recent studies have shown that leukemic clones in approximately 10-20% of CLL patients have stereotypic IgH gene sequences (Stamatopoulos et al., Blood 2007). These stereotypic IgH sequences are characterized by closely homologous complementarity-determining regions 3 (CDR3) sequences. The presence of these sequences supports the model of antigen-mediated disease pathogenesis in CLL. While previous studies have focused on the dominant CLL clone present at diagnosis, recent advances in next-generation sequencing enable analysis of the full IgH repertoire. Here we assessed whether stereotypic IgH sequences are present at very low frequency among the total IgH repertoire in diagnostic and follow-up samples from 36 CLL patients.
Methods:We utilized the LymphoSIGHT method to sequence the IgH repertoire in 36 CLL patients and 185 patients with non-CLL lymphoid neoplasms. Using universal primer sets, we amplified IGH variable (V), diversity (D), and joining (J) gene segments in diagnostic and follow-up samples. Amplified products were sequenced, and CLL-specific clonotypes were identified in the diagnostic sample of each patient based on high-frequency within the B-cell repertoire. We selected a set of 289 stereotypic IgH CDR3 sequences that were previously curated (Stamatopoulos et al., Blood 2007; Messmer et al., Leukemia Res 2008). We assessed whether the stereotypic CDR3 regions were present in the IgH repertoire of both CLL and non-CLL patients using sequence alignment tools.
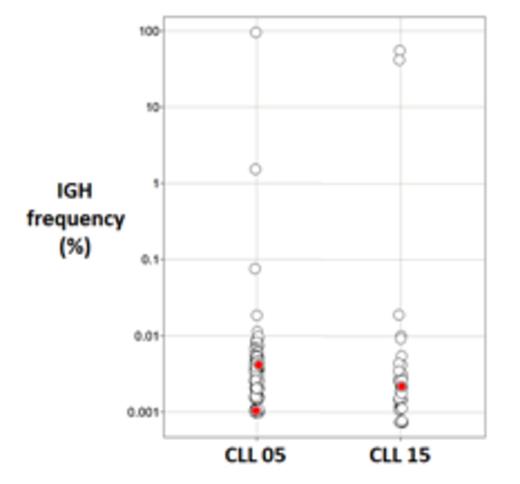
Results: 154,318 IgH clonotypes were observed in the 36 CLL patients, while we identified 8,248,032 IgH clonotypes from 185 patients with non-CLL lymphoid neoplasms. Specifically, we tested 14 mantle cell lymphoma (MCL), 19 hodgkin disease (HD), and 28 acute lymphoblastic leukemia (ALL) patients as well as three cohorts of multiple myeloma (MM) with 26, 30, and 68 patients. We assessed whether each of the 289 CDR3 stereotypes were present in the IgH repertoire of the CLL or non-CLL patients. Twenty two of the stereotypic IgH sequences were observed in any patient: 2 in CLL patients and 20 in the non-CLL patients. This is not surprising given that non-CLL patients have over 50 times the total number of clones compared to the CLL group. One of the stereotypic IgH clonotypes was detected in one CLL patient (p-value= 0.018; not significant with Bonferroni correction). The other stereotypic IgH clonotype was present in two CLL patients but not in any of the non-CLL lymphoid neoplasm cohorts (p=0.00034; 0.0074 Bonferroni corrected). In contrast, we tested for association between each of the 20 stereotypic sequences and each of the 6 control cohorts (3 MM, 1 MCL, 1 HD, and 1 ALL), and only two significant associations were observed (p=0.045 and p=0.01; neither were significant with Bonferroni correction). This underscores that the only significant association was seen in the tested CLL cohort. Of note, both of these stereotypic IgH clonotypes were present at very low frequency in the IgH repertoire (<10-4) (Figure 1). The two stereotypic IgH sequences identified in CLL patients were unmutated and match the previously published V, D, and J segments. Specifically, the stereotypic sequence ARHRLGYCSSTSCYYYYYGMDVWGQGTT present in one CLL patient has IGHV4-39, IGHJ6, and IGHD2-2 and the stereotypic sequence CARDSPLVVPAAIFYYYYGMDVW present in two CLL patients has IGHV3-48, IGHJ6, IGHD2-2. In contrast the vast majority of the hits in the non-CLL samples have V, D, and J sequences that are different from the published segments for the specific stereotypic sequence.
Conclusion : Our results demonstrate that CLL patients harbor stereotypic IgH sequences. These sequences were not the dominant, high-frequency clonotypes identified at diagnosis in the CLL patients and were absent in a much larger set of clones derived from a cohort of non-CLL cancer patients. These results suggest that a subset of CLL patients may possess an antigen environment that selects for multiple B cells with stereotypic IgH sequences at low frequencies. This selection process may predispose patients to CLL that develops later via accumulation of multiple genetic hits in a pre-leukemic cell containing the stereotypic IgH sequence driving it to high-frequency.
Each clonotype observed in two CLL patients (X axis) is shown as a dot. The Y axis depicts the frequency of the clonotypes. The stereotypic clonotypes are shown in red.
Each clonotype observed in two CLL patients (X axis) is shown as a dot. The Y axis depicts the frequency of the clonotypes. The stereotypic clonotypes are shown in red.
Klinger:Sequenta, Inc.: Employment, Equity Ownership. Hervold:Sequenta, Inc.: Employment, Equity Ownership. Faham:Sequenta, Inc.: Employment, Equity Ownership, Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal