Abstract
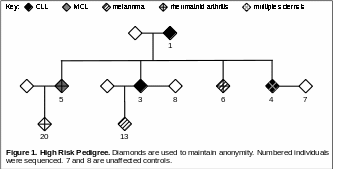
We identified a Caucasian family with dense distribution of B-cell non-Hodgkin lymphoma (NHL) and autoimmune disease (Figure 1). Within the family, three siblings and their mother have B-cell NHL (three with chronic lymphocytic leukemia [CLL]; one with mantle cell lymphoma [MCL]), and four family members have autoimmune diseases (rheumatoid arthritis or multiple sclerosis). In addition, two family members have melanoma. To investigate the genetic factors involved in B-cell NHL in this family, we exome sequenced nine individuals, including two unaffected spouses as technical-artifact controls. Following best-practice guidelines, we performed joint variant calling, including an additional 135 European 1000Genome exomes as a set of background controls.
After variant quality control, a total of 676,137 sequence variants were identified. We also performed individual genotype quality control based on a minimum read depth of 10 and genotype quality of 40, resulting in 554,690 variants. For our initial prioritization, we required that variants be: absent in the spouse controls, have minor allele frequency (MAF) ≤ 0.05 in the 1000Genome exomes, and be shared by all four B-cell NHL cases. This resulted in 73 variants of potential interest.
Of the 73 variants, seven were predicted to be damaging by either PROVEAN or SIFT, two by both algorithms and by PolyPhen2. These two potentially damaging variants reside in the genes CCDC144A and RRS1. The gene CCDC144A is located at chromosome 17p11.2, a known breakpoint region of the 17p deletion often seen in CLL tumor cells. The variant identified in this gene was rare (MAF = 0.001 in the NHLBI Exome Sequencing Project [ESP]) and was also seen in two additional family members (one with rheumatoid arthritis; a second with rheumatoid arthritis and melanoma). The gene RRS1 is located at chromosome 8q13.1, and has been shown to be important for proper chromosomal organization during mitosis. The variant identified in this gene was relatively rare (MAF = 0.041 in ESP) and was seen in the same, two additional family members. Of the remaining five variants predicted damaging by only one algorithm, of interest were two variants in the SGK223 gene on chromosome 8p23.1. SGK223 is a kinase and a component in the BCR-ABL1 signaling network that is present in most chronic myelogenous leukemia cases and a quarter of adult acute lymphoblastic leukemia cases. Both of the variants in SGK223 were also seen in three additional family members (one with rheumatoid arthritis; a second with rheumatoid arthritis and melanoma; a third with melanoma).
In addition to prioritization by predicted function of coding variants, we explored the 73 variants for overlap with findings from published germ-line investigations of B-cell NHL. This identified one variant in the ACOXL gene at chromosome 2q13. The variant is relatively rare (MAF = 0.017 in dbSNP) and lies in the same gene and intron (17,016 base pairs upstream) as rs13401811, the associated SNP (p = 2.08 x 10-18) reported in a genomewide association study (GWAS) for CLL.
Our ongoing analyses and prioritization of the variants in this extraordinary family reveal overlap with signals coming from GWAS studies and suggest some potentially damaging variants in genes not previously implicated in NHL. The potential risk-alleles identified in our preliminary findings could shed new light about the genes and genetic factors involved in NHL.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal