Abstract
Purpose: Chronic Myeloid Leukemia (CML) among other hematological malignancies has witnessed development of advanced molecular diagnostics. However, there is an unmet need for efficient objective markers for selection of available therapeutic agents and accurate prediction of patient’s response. Clinical proteomics can potentially be complementary to currently existing proven tools to monitor therapy response towards achieving personalized medicine for CML patients.
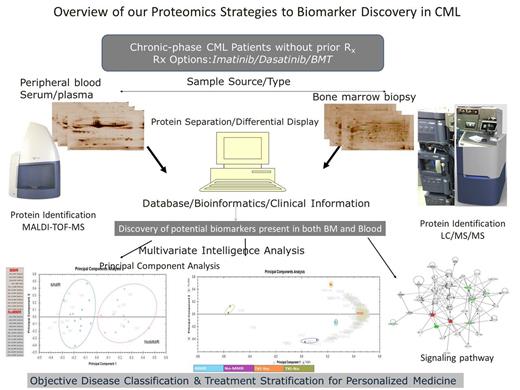
Experimental Design: Thirty four bone marrow and serum, samples from patients with newly diagnosed chronic–phase CML were subjected to expression proteome analysis using combined gel-based 2-DE and label-free in-solution quantitative liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). The study aims to identify disease-specific/disease-associated protein biomarkers demanding less invasive procedures for objective prediction of individual’s best treatment options and prognostic monitoring of CML patients (Overview in figure 1).
Results: Quantitative protein fingerprints from 2-DE analysis accurately predicts 13 individuals that achieved Major Molecular Response (MMR) at 6 months from 12 subjects without Major Molecular Response (NoMMR) using unsupervised principal component analysis (Figure 1 left lower panel). Some of the results were independently validated using label free quantitative liquid chromatography tandem mass spectrometry. Sixty three (63) differentially expressed proteins were identified (> 2- ∞- fold change, p < 0.05).The panel of proteins also discriminates accurately patients that stay on TKI after 1year of Imatinib Rx from patients ultimately requiring alternative treatment (Second Generation TKI/others). We have had more than 2 years follow up of these patients and the same dataset of potential protein biomarkers could still accurately separates all four sample groups into their respective molecular response and treatment sub groups (Figure 1 middle lower panel). Some of the identified proteins were implicated in hematological diseases as potential biomarkers using Ingenuity Pathway Analysis (Figure 1 right lower panel).
Conclusion: Our results highlight the power of Proteomics as a molecular scanner for objective stratification of CML patients for treatment options. We have identified protein signatures capable of prediction of molecular response and choice of therapy for CML patients at 6 months and beyond. Our expression proteomics strategy is very promising for identification of clinically useful biomarkers. These proteins might be valuable once validated, to complement the currently existing parameters for reliable and objective prediction of disease progression, monitoring treatment response and clinical outcome of CML patients as a model of personalized medicine.
Overview of our biomarker discovery proteomics approach. Bone marrow and peripheral blood samples were analyzed by 2-DE and LC/MS/MS. Identified proteins were subjected to multivariate statistical analysis and evaluated for early treatment response and prediction of individualized treatment options. Potential markers would be validated for clinical use.
Overview of our biomarker discovery proteomics approach. Bone marrow and peripheral blood samples were analyzed by 2-DE and LC/MS/MS. Identified proteins were subjected to multivariate statistical analysis and evaluated for early treatment response and prediction of individualized treatment options. Potential markers would be validated for clinical use.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal