Abstract
Background:
Advances in molecular genetics have changed the risk stratification and treatment of patients with Chronic Lymphocytic Leukemia (CLL). Previous studies have shown the worst patient outcomes associated with 17p deletion from diminished overall and progression free survival, in addition to lack of response to conventional Fludarabine based chemotherapy regimens. More recently, analyses of the role of TP53 mutation utilizing next generation sequencing (NGS) in CLL patients has shown that it may also be associated with poor prognosis and similar outcomes to those patients with 17p deletion. This study seeks to characterize the role of TP53 mutation and 17p deletion with overall survival (OS) of CLL patients treated at H. Lee Moffitt Cancer Center who underwent Targeted Exome Sequencing (TES).
Methods:
We utilized the Total Cancer Care/H. Lee Moffitt Cancer Center (MCC) database containing 844 CLL patients of diverse ethnic backgrounds and long survival follow up to determine the rates and outcomes of 17p deletion within our population. A subset of 93 patients treated between 2004 and 2010 at MCC were randomly chosen for TES. Bone marrow and/or peripheral blood samples were subjected to genomic capture and massive parallel sequencing of 1,321 cancer-related genes. Sequences were aligned to the hs37d5 human reference. Insertion/deletion realignment, quality score recalibration, and variant identification were performed with the Genome Analysis ToolKit (GATK). Sequence variants for TP53 and 17p deletion were annotated with ANNOVAR. Alignments using BWA and Stampy were manually inspected with Samtools View. The primary objective was to determine OS stratified into four groups: 1) TP53 positive/17p deletion positive (tp53+/17pdel+), 2) TP53 negative/17p deletion positive (tp53-/17pdel+), 3) TP53 positive/17p deletion negative (tp53+/17pdel-), and lastly 4) TP53 negative/17p deletion negative (tp53-/17pdel-).
Results:
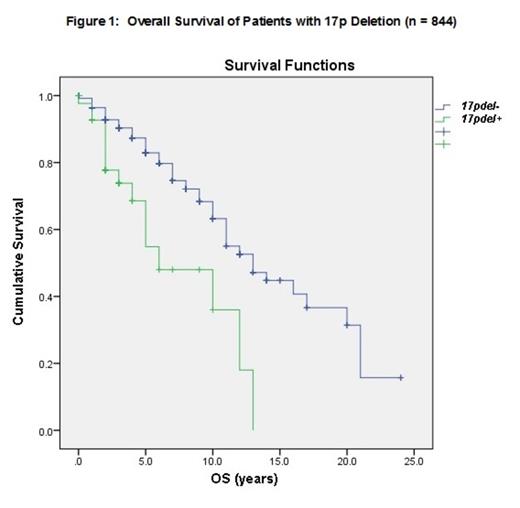
Analysis of patients with genetic data available in our larger population of CLL patients at MCC (n=844), revealed the median OS of patients with 17pdel+ (n=46) was 6 years vs. 13 years for 17pdel- patients (Figure 1, p<0.001). These results were comparable to our CLL patients that underwent TES. Among patients who underwent TES (n=93), the median age was 58 (34-87) years and the male/female ratio was 63/30. Sixteen patients had Rai Stage III/IV disease (17.3%). ZAP70 and CD38 were positive in 41 (44.1%) and 11 patients (11.8%), respectively. Recurrent CLL mutations by FISH showed 13qdel in 54 (58.1%), 11qdel in 19 (20.4%), trisomy 12 in 18 (19.4%), and 17pdel in 12 (12.9%) cases. By TES, TP53 mutation was the most frequent mutation and detected in 18/93 (19.4%) of patients. The median OS for 17pdel+ vs. 17pdel- in this population were 9.6 and 14 years, respectively (p=0.023). When patients were divided by subgroups the frequencies were as follows: tp53+/17p+ in 10 (10.8%), tp53-/17p+ in 2 (2.2%), tp53+/17p- in 8 (8.6%), and tp53-/17p- in 73 (78.5%). The median OS for patients with tp53+/17pdel+, tp53+/17pdel-, and tp53-/17pdel- were 2, 9, and 13 years, respectively (Figure 2, p=0.028). Due to the small number in the tp53-/17pdel+ subgroup, the median OS could not be determined.
Conclusion:
The impact of TP53 mutations detected by NGS in CLL patients is still under investigation. TP53 mutation and 17p deletion are associated with a very poor prognosis. The impact of TP53 mutation in the absence of 17p deletion is not well understood. Within our study, our findings clearly show the 84.6% reduction in OS (11 years) of tp53+/17p+ patients when compared to tp53-/17pdel- patients. TP53 mutation in the absence of 17p deletion did reduce OS by 4 years (30.8%) when compared to patients who lack the 17p deletion or TP53 mutation. TP53 mutation does appear to impact and shorten OS most strikingly in the presence of 17p deletion, suggesting that 17p deletion may play a greater role in prognosis than TP53 alone. Larger number of patients will be needed in order to confirm these findings and to determine the impact of 17p deletion in patients lacking TP53 mutations. Further analyses of CLL patients utilizing NGS technologies and functional analyses to determine if these mutations fully inactivate TP53 will need to be performed to help further elucidate the role of TP53 mutation in patients with high-risk CLL.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal