Abstract
INTRODUCTION: Minimal residual disease (MRD) is defined as the persistence of residual tumor cells in patients during or after treatment, when the patient is in remission. The monitoring of MRD has been shown to be an excellent tool for optimization of patient care and therapy response. Indeed, formal proof of the beneficial effects of MRD measurements has been published for patients with different types of leukemia, and usually quantitative PCR (qPCR) is the methodology employed, as it is currently the most sensitive approach for disease monitoring, though flow cytometry can be a valid alternative. In patients with non-Hodgkin lymphoma (NHL), however, the impact of MRD monitoring is less well defined. Approximately 90 % of MCL patients are diagnosed by means of pathology with overexpression of the cyclin D1 gene (CCND1) caused by the fusion of the BCL1 gene on chromosome 11 to the immunoglobulin heavy chain (IgH) locus on chromosome 14. In addition to t(11;14), clonal rearrangements of the IgH locus can be identified in most MCLs. Recently, the overexpression of the intron-less SOX11 gene was reported as a diagnostic marker highly associated to MCL and has the added advantage that it also identifies the MCL patients negative for CCND1 overexpression.
AIM: Given that the literature is lacking direct comparisons of these four key biomarkers for MRD monitoring we have performed a detailed comparison between these markers.
MATERIALS & METHODS: We employed previously reported assays for sensitive and mRNA specific quantification of the overexpression of SOX11 and CCND1 [Hamborg et al., Eur J Haematol. 2012 Nov;89(5):385-94] as well as the genomic fusion of BCL1/IgH [Sørensen et al., J Immunol Methods. 2014 Apr;406:131-6], by expanding the latter assay with patient-specific forward primers the IgH-VDJ region was targeted. Forty-four diagnostic peripheral blood (PB) samples, 21 relapse PB samples, and 148 follow-up PB samples were analyzed for both SOX11 and CCND1 expression, and a subgroup of these samples was additionally analyzed for BCL1/IgH and clonal IgH-VDJ allelic burden. Part of the patient material has been used in other studies.
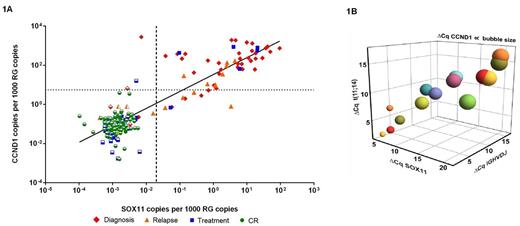
RESULTS: As depicted in Figure 1A, we found a strong correlation (R2 value: 0.7867 – calculated using GraphPad PRISM 6) between SOX11 and CCND1 expression in samples ranging from diagnosis to complete remission (CR) and relapse. While a low background expression was a feature of CCND1 in CR samples we did not observe that for the expression of SOX11 (partly opened symbols in Fig. 1A indicate SOX11 negativity, Cq > 40 and the discontinued lines defines the normal background levels for SOX11 and CCND1 respectively). For the direct comparison between the four markers we selected five representative patients with longitudinal samples available and quantified MRD using all four genetic targets. Fourteen out of 31 samples from these five patients were positive for all four biomarkers (Cq < 40) and were included in the correlation analysis displaying an adjusted correlation of 0.9168 calculated using the Wolfram Mathematica 9 (Fig. 1B, where CCND1 is shown as a function of size). The samples with the highest disease burden showed the lowest ΔCq value, which visually demonstrated the concordance between all markers.
DISCUSSION: Although further studies are required, the present findings suggest that all four biomarkers are suitable for quantification of the molecular targets characterizing the disease burden in MCL. However, the universally expressed CCND1 has a high background expression in patients in CR. While the t(11;14) assay is sensitive, it does not read out in more than half of the patients. The IgH assay, in turn, while also being sensitive is cumbersome to perform with a long turnover time. For SOX11, the assay sensitivity can be limited in patients with low diagnostic expression levels.
CONCLUSION: Taken together, we suggest that at least two of the above markers should be applied to secure that the majority of patients will be amenable MRD for clinical intervention purposes. Based on the data here, we suggest that the combination of SOX11 and t(11;14) is to be preferred, but for patients negative for t(11;14), IgH@ is a valid substitute.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal