Abstract
Background: Acute myeloid leukemia(AML) is a heterogeneous hematological disease characterized by proliferation and accumulation of myeloid precursors in the bone marrow, decrease of apoptosis level and differentiation arrest of these cells. The French, American and British group of leukemia experts (FAB) divided acute myeloid leukemias into eight subtypes based on the characteristics and behavior of leukocytes and the in cell differentiation blockage. Although several studies in the area, events related to the beginning of disease as well as its progression is still unknown. It is believed that malignant transformation in normal Hematopoietic Stem Cells (HSC) can give rise a Leukemic Stem Cell (LSC) and this transformation could be related to changes in Mesenchymal stromal cells (MSC) signaling. In this context, the aim of this work is to analyze the role of MSCs in the leukemic transformation of hematopoietic stem cell. To address this hypothesis we used ChipArray approaches to evaluate the gene expression profile of MSC from AML patients compared to MSC from Healthy donors (HD).
Methods: For this purpose, bone marrow(BM) samples from HD and patients diagnosed with AML previously classified into subtypes were placed in culture. We used 7 samples from different subtypes of AML (four from subtype M2, two from M3 and one from M4/M5) and compared with two pools of HD. After 3rd passage, total RNA from MSCs cultures were obtained using RNeasy Mini Kit according to the manufacturer’s instructions (QIAGEN). After that, RNA was processed according to GeneChip whole transcription (WT) sense target-labeling assay (Affymetrix) and hybridized to GeneChip human gene 1.0 ST array (Affymetrix) washed and stained according to the manufacturer’s protocols. Data were analyzed using Partek® software and differentially expressed genes with ³ 2 fold-change was used as criteria to define overexpression or downregulation.To confirm the array assay we performed real-time PCR with a large number of patients with different subtypes. Pathway analysis and related processes was obtained using the Ingenuity Pathways Analysis (IPA) and MetaCoreTM softwares.
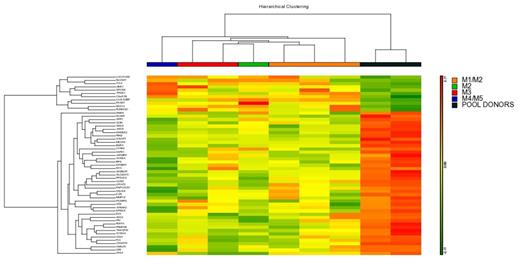
Results: The results obtained in ChipArray assay showed that there is a common molecular signature to all MSCs AML patients compared to donors. Fifty five genes differentially expressed were found. To confirm the results obtained in ChipArray real-time PCR with some genes was performed with more patients (CCL2, SPON2, MUC12 - overexpressed and MMP16, BMP4, CLDN1, SDC2 and OPN downregulate). The results confirmed the commom molecular signature in all AML patients instead of subtypes.These genes are involved in processes of cellular adhesion, differentiation of osteoclast and Wnt pathway. We further carried out another ChipArray assay with another samples. At this time, an unsupervised analysis with the 55 differentially expressed genes was done to verify if the samples agrouping in clusters that separate AML patients from HD. The results confirmed that this molecular signature is capable to distinguish patients from HD. Changes in MSC signaling could be related to malignant transformation of HSC and some of these genes found in ChipArray are secreted proteins that could affect HSC. To verify if these secreted proteins are reallly differentially expressed in AML patiens we performed Western blot assay with plasma samples from AML patients and HD. The secreted proteins seleted were CCL2 and BMP4 (overexpressed and downregulated in AML patients, respectively). The results confirmed the increase of CCL2 protein and decrease of BMP4 protein in AML patients plasma. In Acute Lymphoblastic Leukemia (ALL), CCL2 was able to increase the adhesion of ALL cells to MSC and to promote survival and proliferation of MSC.BMP4 has been described as an important component produced by the hematopoietic microenvironment that regulates both HSC number and function. Moreover, BMP4 has also implicated BMP4 in homing and engraftment of human hematopoietic stem/progenitor cells.
Conclusions: With these results, we could start to elucidate the participation of AML-MSCs in the leukaemogenic process. Altogether, both CCL2 and BMP4 produced and secreted by MSC from BM microenvironment seems to be important for the biology of AML cells.
Financial support: CNPq, FAPERJ, INCT.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal