Abstract
Homeostatic processing of von Willebrand factor (VWF) in the circulation relies on proteolysis of ultra-large VWF (ULVWF) multimers by the metalloprotease ADAMTS13. Deficiency in ADAMTS13 results in Thrombotic Thrombocytopenic Purpura (TTP) due to the accumulation of thrombogenic ULVWF. ADAMTS13 is secreted into the circulation in its active form and has no known inhibitor. Regulation of ADAMTS13 activity occurs in part through plasma ADAMTS13 levels and by the hemodynamic exposure of the scissile bond in VWF.
To identify the genetic determinants of ADAMTS13 levels we performed genome-wide association studies (GWAS) and linkage analysis in two independent cohorts of young healthy individuals. This study recruited 3206 young healthy individuals of European descent who participated in the Genes and Blood Clotting Study (GABC, n=940) and the Trinity Student Study (TSS, n=2266). All subjects provided written informed consent and all genotyping and phenotyping was performed on de-identified samples. Details of the genotyping and data cleaning process for these cohorts were previously described. (Desch KC et al. PNAS 2013, 110:588-593) ADAMTS13 levels were measured by a custom AlphaLISA (Perkin-Elmer, Waltham, MA) assay using polyclonal anti-ADAMTS13 antibodies (a gift from X. Long Zheng, University of Pennsylvania).
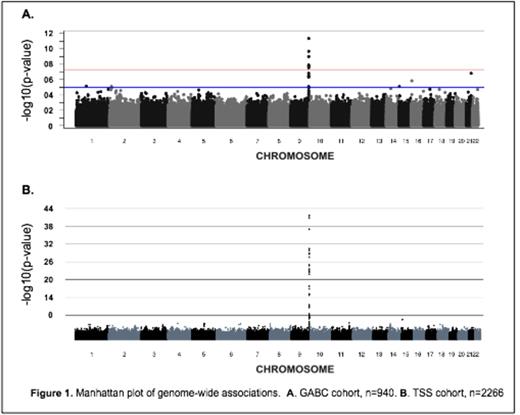
In GABC, the median ADAMTS13 levels were 115.4 IU/dL and demonstrated a 2.0-fold variation between the 5th and 95th percentiles. Heritability of ADAMTS13 levels was estimated at 77% in GABC and 43% in the TSS. ADAMTS13 levels were associated smoking in both cohorts and was associated with an 8.9% increase in levels of ADAMTS13 (P < 1.5E-11, GABC). GWAS identified 10 SNPs in GABC and 33 SNPs in TSS that were significantly associated with ADAMTS13 levels (P<1.0E-8), Figure 1 . In both cohorts, significant signals were located at the ADAMTS13 gene on chromosome 9q34. The top SNP association signal in GABC was at rs2073933 (P < 4.48E-12) and the minor allele was associated with a 6.9% decrease in ADAMTS13 levels. In the TSS study this SNP was also significant (P < 2.37E-42) and associated with a 5.2% decrease in ADAMTS13 levels. In both GWAS studies, no SNPs remained significant after the top ADAMTS13 SNP was used as a covariate. Linkage analysis, which maps genomic regions where allele sharing patterns in families correlate to ADAMTS13 levels, identified 2 regions with LOD scores greater than 3. The first signal was on chromosome 9q34, including ADAMTS13 (LOD 4.12) and the second was on chromosome 5p13 including EGFLAM (LOD 3.08).
Although ABO mediated glycosylation patterns affect VWF clearance rates, no signal in ABO was detected in our multiple association studies suggesting that the common ABO haplotypes are not associated with ADAMTS13 levels. Although other mutations tagged by the significantly associated SNPs cannot be excluded, a potential candidate mutation (rs2301612, Q448E) was identified in ADAMTS13 and may be responsible for the decrease in ADAMTS13 levels through alteration of synthesis/secretion or plasma clearance rates. Aditionally, linkage analysis confirmed the chromosome 9q34 signal and identified a region on chromosome 5p13 that was not detected by GWAS. This finding suggests that rare variants in this genomic region also contribute to ADAMTS13 levels. The identification of mutations that determine normal ADAMTS13 variation may lead to a better understanding of the genetic risk for TTP and mechanisms of von Willebrand Factor homeostasis.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal