Abstract
BACKGROUND: Mean life expectancy among adults with sickle cell disease (SCD) remains in the sixth decade of life (Elmariah et al. Am J Hem. 2014). We examined the relationship of genetic differences among SCD patients to survival, in order to gain greater understanding of contributors to early mortality. Genetic signatures associated with survival could identify high risk patients who would be optimal candidates for advanced therapies.
METHODS: Patient data were previously collected through a multicenter study to identify genetic factors associated with SCD complications. All enrolled patients were 18 years or older, with a diagnosis of SCD (SS, Sβ0, Sβ+, SC). Of the patients enrolled, 542 were followed for a mean of 9.3 years for incidence of death. Genome-wide association study (GWAS) data from the Illumina 610 BeadChips were available for 503 of these individuals. We used the software program IMPUTE2 with a global reference panel from the 1000Genomes Project to impute missing genotypes up to ~3 million single nucleotide polymorphisms (SNPs). Time to death was correlated with genotype using a Cox Proportional Hazards model implemented in an R plugin to PLINK (Purcell et al. AJHG. 2007), controlling for age at enrollment, gender, and population substructure. Candidate genes were prioritized for follow-up using the following criteria: at least one SNP with p<0.00001, additional significant SNPs with p<0.0001 within the same gene, and biological plausibility. To confirm the associations observed with the most significant imputed SNPs, follow-up genotyping was performed by TaqMan analysis (N=15 SNPs). Post-hoc regression analysis of all available SNPs in the five candidate genes (N=2549) and clinical phenotypes previously associated with survival (Elmariah et al. Am J Hem. 2014) was performed in PLINK. False discovery rate p-values were obtained with PROC MULTTEST in SAS version 9.4 (SAS Systems, Cary, NC).
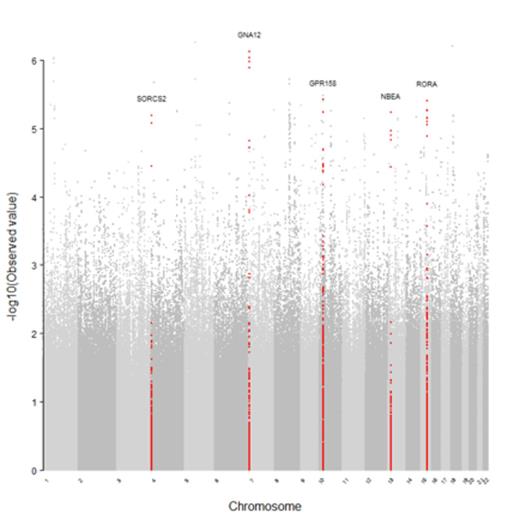
RESULTS: Median survival was 61 years for all subjects. Median survival for patients with hemoglobin (Hb) SS and Sβ0 was 58 years and for Hb Sβ+ and SC was 66 years. GWAS results are shown in FIGURE 1. No genome-wide associations with survival met adjustment for multiple testing. The hazard ratios for the most significant SNPs increased risk for mortality 2-3 fold with each minor allele present. Five candidate genes were selected for follow-up: G-Protein Coupled Receptor 158 (GPR158), G protein alpha-12 (GNA12), Neurobeachin (NBEA), RAR-related orphan receptor alpha (RORA), and Sortilin-related VPS10 Domain Controlling Receptor (SORCS2). Follow-up genotyping of fifteen selected imputed SNPs within the candidate genes confirmed the imputed genotypes and associations from GWAS (p's<0.00001). Several SNPs in GPR158, representing one linkage disequilibrium (LD) block, were associated with seizures (FDR p<0.05, OR=2.1). Likewise, several SNPs in GNA12, representing one LD block, were associated with glomerular filtration rate (GFR; p<0.001, OR=-24.6) but did not reach FDR significance.
CONCLUSIONS: We have identified genes that were nominally associated with survival in our SCD cohort and had clinical relevance. GNA12 has been associated with cellular adhesion, proteinuria, and renal failure (Boucher et al. Lab Invest. 2012). We previously linked kidney disease to early mortality in SCD (Elmariah et al. Am J Hem. 2014)and confirm here that GNA12 is associated with GFR and survival. In our cohort, GPR158 is associated with seizures, and other candidate genes, including NBEA, RORA, and SORC2, have previously been associated with neurologic development and function. These findings corroborate our prior results that seizures and strokes are strongly associated with decreased survival in SCD patients. The association between RORA and survival might be attributable to its role in nitric oxide biosynthesis. Several nominally significant SNPs lie in known regulatory regions, which may explain mechanisms by which those SNPs influence mortality. Our study does not prove causality or confirm mechanism of action. Further, we had limited statistical power due to the relatively small cohort, evidenced by the lack of significance on false discovery rate testing. Despite these limitations, our results demonstrate that genetic signatures may identify high-risk SCD patients who might benefit from more aggressive novel therapies and provide interesting mechanistic hypotheses.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal