Abstract
Tet2 catalyzes the conversion of 5-methylcytosine to 5-hydroxylmethylcytosine (5hmC), which is considered the first step in active DNA demethylation. TET2 is frequently mutated in various hematopoietic malignancies. Mutations in TET2 and DNMT3A often co-occur in T cell lymphoma patients, though they work in the same DNA methylation-hydroxymethylation pathway. Here, we explored whether TET2 and DNMT3A could cooperate and how they interact in malignant hematopoiesis using an Mx1-cre+; Dnmt3af/f; Tet2-/- (DKO) mouse model, using as controls Mx1-cre; Dnmt3af/f (Dnmt3a KO) mice, Tet2-/- (Tet2 KO) mice and WT mice.
We performed whole bone marrow transplantation of each genotype in competition with WT bone marrow to compare the competiveness of mutant stem cells. We found the order of engraftment activity to be: DKO > Tet2 KO > Dnmt3a KO, suggesting Dnmt3a loss-of-function augments the competitive advantage of Tet2 KO cells. Consistent with our results, DKO recipients showed a threefold increase in Lin-Sca1+cKit+ (LSK) population compared to Tet2 KO recipients 6 months after transplantation. There was also an increase in short-term hematopoietic stem cells, showing Dnmt3a loss can increase the stem cell and progenitor pool in Tet2 KO background. Forty weeks after transplantation, DKO recipients developed lethal B cell ALL characterized by leukocytosis and anemia, while Tet2 KO mice did not show signs of hematopoietic malignancy one year after transplantation.
We then isolated genomic DNA and RNA from Tet2 KO and DKO long-term hematopoietic stem cells (Lin- cKit+Sca1+ CD48- CD150+) and performed whole genome bisulfite sequencing and RNA-seq. We compared the transcriptome and methylome of DKO, Tet2 KO, and Dnmt3a KO with that of WT HSCs, and we identified the differentially expressed transcripts and DMRs in the DKO that overlap with those in the single mutants, as well as those unique to the DKO. At the transcriptome level, the DKO HSCs overexpressed multiple transcription factors associated with differentiated lineage - lymphoid Ikzf1, Pax5, and Ebf1, myeloid Cebpa and Cebpe, and erythroid Klf1. Specifically, overexpression of the erythroid gene signature controlled by Klf1 was observed only in DKO HSCs. This signature is also found in AML patients carrying both DNMT3A and TET2 mutations, suggesting Klf1 overexpression in DKO HSCs could be the driving event of leukemogenesis. Further, we found the expression levels of Klf1 and Ikzf1 across the genotype in following order: DKO>Tet2 KO>WT>Dnmt3a KO, suggesting Tet2 represses these genes in wild type HSCs.
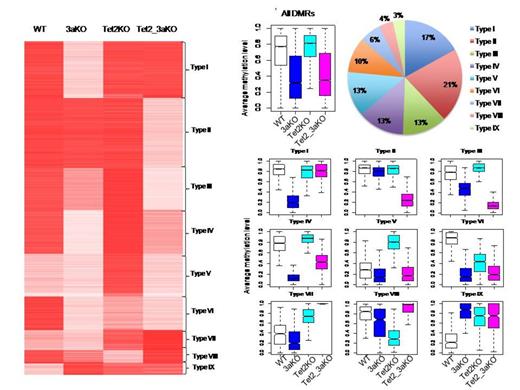
At the methylome level, most hypomethylated differential methylated region (DMR) in DKO HSCs are already hypomethylated in Dnmt3a KO HSCs - evidence that Dnmt3a is upstream of Tet2 in the regulation of the methylome. We identified 9 major dynamic methylation patterns across the 3 genotypes, including one that indicates that Dnmt3a and Tet2 are counteractive. However, in about 15% of these DMRs, methylation levels in the DKO decreased to an average of 10%, while the same genomic regions of the Dnmt3a KO had average methylation levels of 50% (a 30% decrease compared with WT). This significant decrease in Dnmt3a KO indicates that Tet2 maintains the methylation at these DMRs in Dnmt3a KO HSCs. Further investigation shows that these DMRs are enriched in 5hmC in the Dnmt3a KO HSCs, which implies that Tet2 is hydroxymethylating the DMRs at these sites when Dnmt3a is lost.
Both Klf1 and Ikzf1 harbor a DMR of this kind in their upstream promoter regions. The hypomethylated DMR associated with Klf1 is already hypomethylated in the Dnmt3a KO HSCs, but the methylation level further decreased in the DKO from 50% (Dnmt3a KO) to 0.02% (DKO). Elevated 5hmC is also observed in the DMRs of Klf1 and Ikzf1 in Dnmt3a KO HSCs. 5hmC is thought to be associated with both repressive and activating marks, and the expression of Klf1 and Ikzf1 is repressed in Dnmt3a KO HSCs but upregulated in Tet2 KO. Thus we propose that DNA methylation at regulatory regions of these factors decreases in Dnmt3a KO HSCs, but Tet2 represses their expression by hydroxylmethylation. The loss of both Tet2 and Dnmt3a is required for full activation of Klf1 and Ikzf1.
Therefore, we conclude that in HSCs both Tet2 and Dnmt3a likely repress differentiation-associated transcriptional factors by hydroxymethylating and methylating at regulatory regions.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal