Abstract
Background: The invention of Next Generation Sequencing (NGS) has spurred research into human diseases, especially in the field of malignancy. In acute myeloid leukemia (AML), a plethora of novel alterations have been identified, including mutations in epigenetic regulator genes (e.g. IDH1, IDH2, DNMT3A), genes coding for proteins of the cohesin complex (e.g. SMC1A, SMC3, STAG2) and spliceosome genes (e.g. SF3B1, U2AF1). Although the diagnostic and prognostic implications of many of these alterations are not yet clear, there is increasing evidence that several of them might have major implications for understanding the disease biology or for patient-treatment. Thus, there is increasing need to reliably detect these mutations in large patient groups in clinically relevant time-frames and at an affordable cost. Due to the large number of genes to be screened, amplicon-based NGS represents an attractive detection method. Although, several assays have been reported, integrating different numbers of genes, it is currently unclear whether they really allow reliable detection of alterations in a reproducible way. Here we report our results from a round robin comparison of the detection of known AML-variants using a highly multiplexed, single tube assay coamplifying a total of 568 amplicons covering 54 entire genes or hot spot gene regions involved in leukemia (TruSight Myeloid sequencing panel; Illumina), with respect to the sensitivity, reproducibility and quantitative accuracy.
Material and Methods: Ten European laboratories routinely involved in molecular AML diagnostics participated in this study. All groups performed two sequencing runs, each containing 8 samples. These samples were centrally aliquoted and distributed, the analyses were done in a blinded fashion. Six out of the 8 samples on each run were derived from a set of 9 samples composed of DNA isolated from the blasts of 18 different newly diagnosed AML patients mixed at a 1:1 ratio, with 50 ng of DNA being used for the library preparation. Three of these 9 samples were analyzed in replicate in separate runs by each group. The remaining two samples were a commercial test DNA containing 10 known single nucleotide variants (SNV) or insertion/deletion (InDel) alterations with defined variant allele frequencies (VAF) between 4 and 25% and DNA derived from the OCI-AML3 AML cell line (mutant for DNMT3A and NPM1). Sequencing was performed on MiSeq NGS systems (Illumina) using 2x151 bp-runs. Sequencing data were analyzed by all laboratories using the VariantStudio software (Illumina), with the threshold for mutation calling set at 3%.
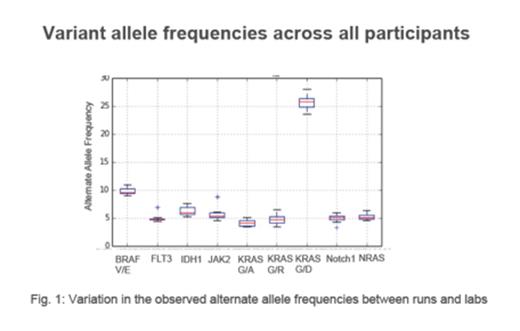
Results: Analysis of data quality indicated that 85% of the samples met the predefined acceptance criteria (>=95% amplicons with at least 500 reads/amplicon), the median coverage was 7379 reads/amplicon (range 0-47403 reads). Of the 9 mutations present in the positive control, 7 were called at least once in the two replicates by all labs, two mutations with a VAF of 5% were missed by 1 and 4 participants, respectively. Overall, the VAF calls for this sample showed a high level of accuracy across the participants (median coefficient of variation 5%, range 0-22.5%) as well as excellent intra- and inter-laboratory reproducibility (Fig.1). In total, the 9 primary leukemic samples contained 43 known variants in 19 genes, including all commonly mutated genes in AML, i.e. CEBPA, DNMT3A,RUNX1, NPM1, FLT3, WT1. For these samples, the sensitivity was 95.7%. Based on the entire data set (positive control and leukemic samples), the calculated sensitivity of the assay for known variants with an expected VAF>=5% was 93.3%. The rate of non-calls was slightly higher for InDels (14/179; 7.8%) than for SNVs (25/407; 6.1%; P=.47). Two 57-bp long insertions in FLT3 exons 14/15 were not called, which is expected due to the specifications of the assay (max. detectable InDel length <=25 bps). The standard deviation of VAF estimates for the primary leukemic samples was 1.7% with a mean CV of 0.094.
Conclusions: This inter-laboratory comparison shows a high sensitivity and impressive quantitative accuracy of NGS-based characterization for known variants down to a minor VAF of 5%. Although additional optimization in individual parameters might be necessary, these initial results clearly indicate that rapid comprehensive molecular characterization of patient samples appears feasible, even in the clinical setting.
Thiede:AgenDix GmbH: Equity Ownership, Research Funding; Illumina: Research Support, Research Support Other. Bullinger:Illumina: Research Support Other. Hernández-Rivas:Illumina: Research Support Other. Heuser:Illumina: Research Support Other. Preudhomme:Illumina: Research Support Other. Lo Coco:Illumina: Research Support Other. Martinelli:Illumina: Research Support Other. Schuh:Illumina: Research Support Other. Enjuanes:Illumina: Research Support Other. Lea:Illumina: Research Support Other. Schlesinger:Illumina: Employment.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal