Abstract
Introduction
Transcriptome analysis by next-generation sequencing (RNA-seq) allows investigation of hematologic malignancies at unsurpassed resolution and provides promising insights into their molecular etiology. Dissecting the underlying biology depends on specific molecular signatures deregulated in the disease subtypes. In addition, gene expression profiles may potentially be used to identify driver genetic alterations and stratify patients based on molecular subtype. In this study, we generated gene expression profiles from RNA-seq data derived from patients with hematologic disease, including acute myeloid leukemia (AML), acute lymphocytic leukemia (ALL), chronic myelomonocytic leukemia (CMML), myelodysplastic syndrome (MDS), chronic myeloid leukemia (CML) and multiple myeloma (MM). Using these data, we aimed to identify disease-specific differentially expressed genes and gain better understanding of the biological functions of these genes for the development of biomarkers and therapeutic strategies in these malignancies.
Methods
Mononuclear cells (MNCs) were isolated from bone marrow or peripheral blood by Ficoll separation from AML (n=24), ALL (n=5), CMML (n=10), MDS (n=4), CML (n=2) and MM (n=7) patients. For MM, CD138+ cells were enriched from MNCs by immunomagnetic bead selection. Total RNA from isolated cells was depleted of ribosomal RNA and reverse transcribed for cDNA. RNA-seq libraries were prepared and sequenced on the Illumina HiSeq instrument. Reads were filtered and aligned to the hg19 human reference genome using TopHat. Second step normalization was carried out to compare gene expression values (FPKM) across samples using a normalization factor derived from 18 reference genes. A log normalized relative expression value for each gene was calculated compared to the median value across all samples. Average linkage based hierarchical clustering was performed and then visualized with TreeView. Network and pathway analyses were performed with IPA (www.ingenuity.com) and Cytoscape¨.
Results
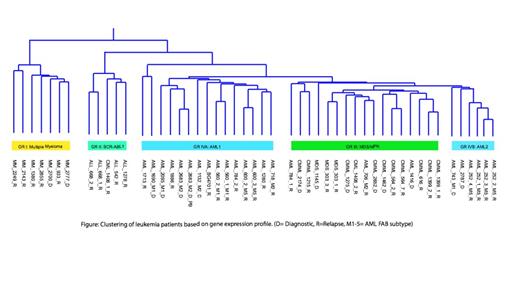
Unsupervised hierarchical clustering of all samples resulted in grouping based on clinical phenotypes with a unique gene signature characteristic for each group (Figure). Analyses of different hematologic malignancies ensured credibility of the classification and highlighted differences in underlying cell signaling networks of each disease. Group I consisted of the multiple myeloma samples, where we identified 25 frequently upregulated genes. Network analyses revealed expression of the upregulated molecules is controlled by two major transcription factors, IRF4 and JUN, which represent the major hubs of the gene signature network. Group II consisted of samples with the BCR-ABL1 fusion and BCR-ABL-like ALLs. Enriched genes in this subgroup included regulators of B-cell development and maturation, plus genes involved in antigen presentation including TCL1A, CD19, CD79, HLA-DQA1, HLA-DQB1 and HLA-DRB1. Group III represented myelodysplastic and myeloproliferative neoplasms and included the MDS and CMML samples. A set of 14 genes differentially enriched in this group formed a unique pro-inflammatory signature. This included TNF-α and IL1B, which act as major regulators of smoldering inflammation driving NF-κB activity and orchestrating downstream activation of signature genes. While MM is known to have activated NF-κB signaling, the gene expression signatures of the MM and MDS/MPN groups were distinct from each other, and included activation of separate sets of cytokines and chemokines. AML samples exhibited heterogeneity in gene expression and formed two groups (IVA, IVB). A HOX gene family expression signature was observed in the FLT3-ITD positive AML samples.
Summary
Our results show that RNA-seq can be used to identify dominant gene expression patterns characterizing different hematologic disease samples, including those sharing a common genetic base (e.g. BCR-ABL, FLT3-ITD) or clinical phenotype (e.g. MDS/MPN, MM). Based on our results, IRF4 may be an attractive therapeutic target for MM. CMML is difficult to diagnose, however, it can be defined by a set of differentially expressed genes that could potentially be used as diagnostic markers. We also show a pivotal role for NF-κB and TNF-α signaling in the pathogenesis of MDS/MPN suggesting that drugs targeting these factors may be useful for the treatment of these diseases.
Porkka:BMS: Honoraria; BMS: Research Funding; Novartis: Honoraria; Novartis: Research Funding; Pfizer: Research Funding. Kallioniemi:Medisapiens: Consultancy, Membership on an entity's Board of Directors or advisory committees. Heckman:Celgene: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal